Accueil du site > Thèmes de recherche > Previously Hosted Projects > Organisation du génome et structuration de la chromatine - Genome Organization and Chromatin Structure > Présentation
Présentation
par Webmaster - 19 février 2013

L’équipe s’intéresse à la structure et la dynamique des macromolécules biologiques ; elle regroupe des compétences de la physique théorique à la bioinformatique.
Dans une étroite relation avec l’expérimentation, les activités de l’équipe visent à l’analyse de la structuration du génome dans les cellules en connexion avec la transcription et la réplication.
La spécificité de l’équipe est d’aller de l’analyse des séquences d’ADN avec des méthodologies originales du traitement du signal (transformation en ondelettes) à la modélisation inspirée de la théorie des systèmes dynamiques (modes normaux, dynamique moléculaire) et de la mécanique statistique (physique des polymères).
The "Genome organization and chromatin structure" group is composed of theoretical physicists with expertise in the study of multi-scale phenomena (turbulence, crystal growth, financial time series, ...) combining the use of concepts from dynamical systems theory and statistical physics with the development of multi-resolution signal and image processing techniques (wavelet transform).
For about a decade, our group has been extending its field of research to the study and modeling of the structure and dynamics of biological molecules (DNA, proteins). In a first step, when using our wavelet-based methodology to analyze the scale-invariance properties of DNA walk profiles generated with some structural tri-nucleotide codings of DNA sequences, we have revealed the existence of long-range correlations (LRC), up to 40kbp, in the fluctuations of the double helix local curvature. These LRC are the signature of the DNA-histone proteins interactions within the nucleosomes, the basic units for DNA compaction in eukaryotic cell nuclei, that constitute a regulatory factor for accessibility to genetic material. When further modeling DNA as a semifexible polymer explicitly taking into account the structural disorder induced by the sequence, we have provided some evidence that the observed LRC favor the spontaneous formation as well as the cooperative positioning of nucleosomes along the chromosomes including the nucleosome free regions experimentally observed at gene promoters (see figure below).
Recently, we have broaden our studies of genomic sequences to encompass scales of the order of chromosome lengths (Mbp) while considering alternative codings with a clear functional significance. During evolution, DNA transcription and replication induce some compositional asymmetry (skew) along the DNA sequences. Deploying a multi-scale strategy of sharp upwardjump detection in noisy skew profiles, we have identified more than 1000 putative master origins of replications that are central to the Human genome organization. Around these putative origins, genes are abundant and broadly expressed, and their transcription is co-oriented with replication fork progression. These features weaken progressively with the distance from putative replication origins. We have proposed that this specific organization could result from the constraints of accommodating the replication and transcription initiation processes at chromatin level, and reducing head-on collisions between the two machineries. Our findings has provided a new model of gene organization in the human genome, which integrates transcription, replication, and chromatin structure as coordinated determinants of genome architecture.
Team Leader ( Chef d’Equipe ) :
Team Members ( Membres de l’Equipe ) :
- Benjamin AUDIT - CNRS Research Associate (1st class)
- Guillaume CHEVEREAU - PhD student
- Cédric VAILLANT - CNRS Research Associate (1st class)
- Lamia ZAGHLOUL - PhD student
- Antoine BAKER - PhD student
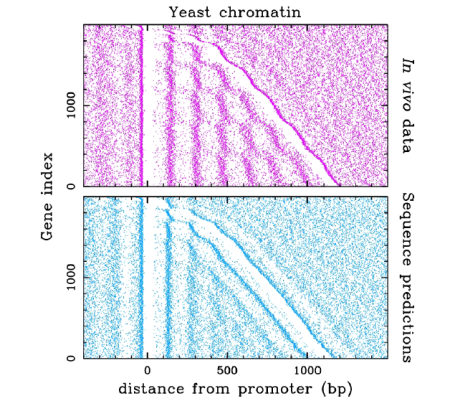
Nucleosome positioning via excluding genomic energy barriers.
Nucleosome occupancy (white corresponding to large occupancy values) for the 2000 shortest S. Cerevisiae genes ordered by increasing length from top to bottom and aligned on the promoter (null distance).
(top) Tiled microarray experimental data (Lee et al., Nature Genetics 39, 1235 (2007)).
(bottom) Predictions based on a physical modeling of sequence-dependent nucleosome energy profile.
Note that the organization of the nucleosomal assembly results from the confinement of nucleosomes between sequence induced high energy barriers corresponding to the nucleosome free regions observed at the promoters and gene ends.
Sélection de publications :
- B. Audit, S. Nicolay, M. Huvet, M. Touchon, Y. D’aubenton-carafa, C. Thermes & A. Arneodo. DNA replication timing data corroborate in silico human replication origin predictions. Phys. Rev. Lett. 99, 248102 (2007).
- M. Huvet, S. Nicolay, M. Touchon, B. Audit, Y. D’aubenton-carafa, A. Arneodo & C. Thermes. Human gene organization driven by the coordination of replication and transcription. Genome Res. 17, 1278-85 (2007).
- C. Vaillant, B. Audit & A. Arneodo. Experiments confirm the influence of genome long-range correlations on nucleosome positioning. Phys. Rev. Lett. 99, 218103 (2007).