Research
RESEARCH AXES
- Chromosome organization & dynamics [PI: D. Jost]:
The chromosome architecture inside cell nuclei plays important roles in regulating cell functions. There are now proliferations of experimental studies which report the hierarchical organization of chromosomes and how this non-random folding plays a role in gene regulation. However, the mechanistic principles that drives such higher-order organization are not fully understood. In close collaboration with experimental biologists, we are developing quantitative polymer models in various organisms to better understand the coupling between 3D chromosome organization and dynamics and biological functions.
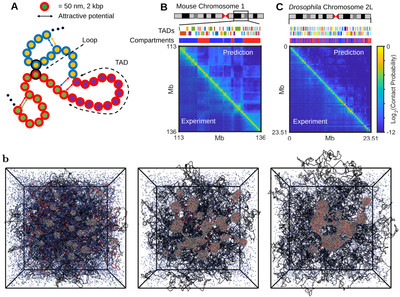 Fig.1: Polymer models and simulations of chromosome organization
Fig.1: Polymer models and simulations of chromosome organization
- Epigenetic (de)regulation & Health [PI: D. Jost]:
Accessibility of transcription factors to promoter is controlled by biochemical modifications of DNA or histone tails which modulate gene expression without alteration of the genetic information. Propagation, maintenance and inheritance of these epigenetic marks are crucial mechanisms in development, cellular homeostasis and disease. With molecular biologists and clinicians, we are developing computational methods and mechanistic models to better characterize epigenetic regulation in normal or cancerous cells and the coupling with metabolism.
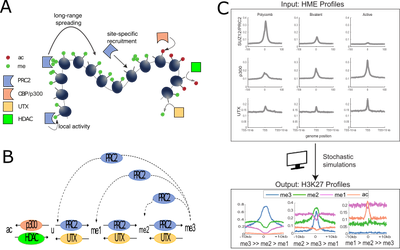 Fig.2: Stochastic models and simulations of epigenomic regulation
Fig.2: Stochastic models and simulations of epigenomic regulation
-
Replication initiation [PI: J-M Arbona]:
Before a cell can divide, it must duplicate its DNA. DNA replication starts at specific sites in the genome called replication origins. Proper regulation of these origins and their timing of firing is fundamental for genome homeostasis. In collaboration with experimentalists, we are interested in the molecular mechanisms of replication initiation and in particular in the mechanism of diffusion of replication activating factors in eukaryotes. We are developing biophysical models and data analysis tools to address these mechanisms and their biological determinants.
-
Deep learning for biology [PI: J-M Arbona]:
In the recent years, deep learning became an essential tool in computational biology. In direct relation to our research, we are also developing various neural network tools for pattern discovery in nanopore sequencing electric signals and in epigenetic profiles. In collaboration with theoreticians, we aim at improving the interoperability of patterns discovered by neural networks.
Funding
Work in the lab is funded by Agence National de la Recherche, CNRS, ENS de Lyon, IXXI.




Networks
The lab is part of several national and international networks: GDR ADN&G, GDRI EpiGeneSys3, COST International Nucleome Consortium.



