Comparing organ development with single-cell and bulk transcriptomes
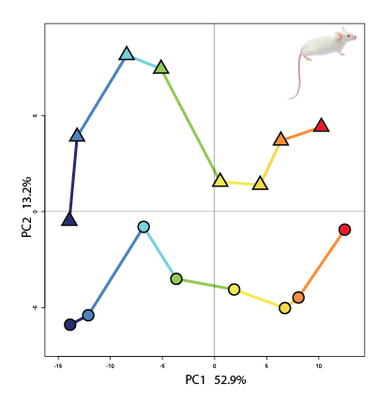 |
Principal component analysis is a useful tool to compare transcriptome timeseries |
Comparing transcriptomes of different molar germs (lower/upper molar), different molar parts (buccal/lingual), and single cells in different species yield insights in the nature and abundance of cell populations throughout development. These are precious informations to understand how development results in different final morphologies and differs between species.
We also ask if the genes expressed during tooth development follow a hourglass conservation pattern as seen with whole body embryonic development.
See our paper in Genome Biology and the accompagnying dispatch by Maayan Baron and Itai Yanai. See also a short advertisement from the CNRS/INSB webpage (in french). See also our latest preprint.
We are taking an active part in the "Spatial Cell ID" Equipex and collaborate on transcriptome analysis (see here and here).
People involved in the team:
Present: Claudine Corneloup
Past: Jeremy Ganovsky, Mathilde Estevez-Villar, Marion Mouginot, Manon Peltier, Alain Rubod, Anne Lambert.
Collaborations: Laurent Guéguen, Université Lyon 1; Marc Robinson-Rechavi, UNIL, Switzerland; Michaelis Averof and Mathilde Paris, IGFL, Lyon; Carina Mugal, Sweden.
