Contrôle de l'expression des gènes par les hélicases à ARN DDX5 et DDX17 - Cyril Bourgeois
Contrôle de l'expression des gènes par les hélicases à ARN DDX5 et DDX17
 Responsable de projet : Cyril BOURGEOIS (cyril.bourgeois@inserm.fr)
Responsable de projet : Cyril BOURGEOIS (cyril.bourgeois@inserm.fr)
ResearcherID E-6337-2010
ORCID 0000-0002-0756-5501
Loop
LinkedIn
Twitter @C_Bourgeois_ENS
DDX17 et DDX5 sont deux hélicases à ARN qui exercent différentes fonctions moléculaires contribuant au contrôle de l'expression des gènes. Elles sont notamment impliquées directement dans plusieurs mécanismes généraux de maturation des ARN (épissage des ARNs pré-messagers, maturation des précurseurs de microRNAs), et nos travaux ont contribué à mettre en évidence leur rôle important dans le contrôle de l'épissage alternatif (1,2,4). DDX17 et DDX5 contrôlent aussi directement la fixation et l'activité de nombreux facteurs de transcription au niveau de la chromatine, par des mécanismes encore mal connus (4,5). Ainsi, l'action à plusieurs niveaux de ces deux hélicases permet une orchestration dynamique de l'expression des gènes qui est fondamentale lors de certains processus de différenciation cellulaire (1-4). Nous avons par exemple montré le rôle essentiel de DDX17 et DDX5 au cours de la différenciation myogénique ou de la transition épithélio-mésenchymateuse (1), une transformation cellulaire survenant lors du développement embryonnaire mais aussi au cours des phénomènes métastatiques. Plus récemment nous avons également mis en évidence un rôle de DDX17 dans le contrôle des phases précoces de différenciation des cellules de neuroblastome (4).
Nous nous intéressons aux mécanismes chromatiniens impliquant DDX17 et DDX5 et régulant le processus transcriptionnel dans son ensemble, c'est-à-dire la transcription proprement dite mais aussi les étapes co-transcriptionnelles telles que l'épissage ou la polyadénylation. Nous avons récemment montré que DDX17 et DDX5 influencent le repliement spatial des gènes et que cette organisation en 3D a un impact sur l'expression génique dans des cellules humaines (6). Outre des techniques traditionnelles de biologie cellulaire et moléculaire (transfection de cellules, analyse d'expression d'ARN et de protéines…), nous utilisons des approches plus exploratoires permettant des modifications ciblées du génome (CRISPR-Cas9) ou l'analyse de l'organisation 3D des gènes (3C/4C). Nos recherches s'appuient beaucoup sur l'analyse de données NGS (next generation sequencing) par des approches computationnelles permettant une vision de ces mécanismes à plus large échelle (RNA-seq, ChIP-seq…).
Dans la continuité de nos travaux précédents, nos objectifs sont de mieux comprendre au niveau moléculaire comment DDX5 et DDX17 régulent l'expression des gènes et la maturation des ARN pré-messagers. Les projets de mon groupe s'articulent autour de 2 axes principaux liés à deux types de pathologies différentes ayant comme point commun une altération de l'expression ou de la fonction de DDX17: un cancer (le neuroblastome) et une maladie génétique (un syndrome neurodéveloppemental associé à des mutations du gène DDX17).
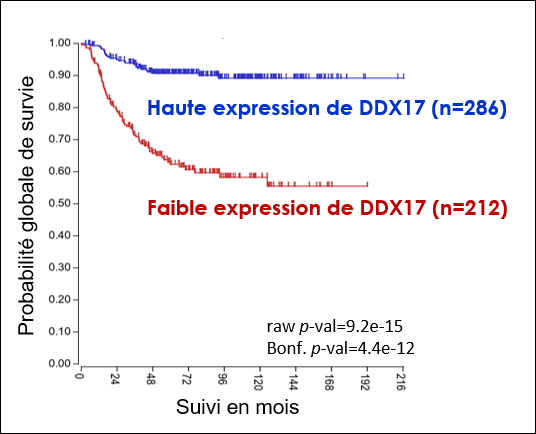
1. Le neuroblastome est un cancer touchant les jeunes enfants et qui reste souvent incurable dans ses formes les plus sévères. Un aspect essentiel du développement plus ou moins agressif des neuroblastomes réside dans la capacité des cellules du système nerveux périphérique à proliférer de façon aberrante ou au contraire à se différencier en cellules nerveuses bénignes, un processus qu'il est très difficile de contrôler thérapeutiquement. DDX17 est impliquée dans la différenciation des cellules de neuroblastome (4), et son expression est de bon facteur pronostic pour ce cancer. Ces résultats suggèrent qu'une expression réduite de ces facteurs, qui entraîne in cellulo de profondes altérations transcriptomiques (non génomiques), pourrait contribuer au développement des neuroblastomes les plus agressifs, une hypothèse que nous souhaitons explorer.
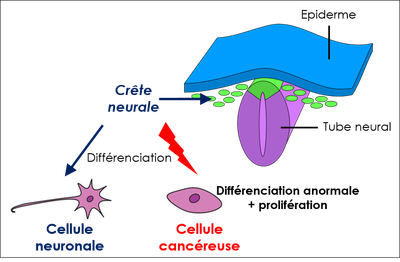
2. Impact de mutations du gène DDX17 dans un syndrome neurodéveloppemental. En collaboration avec le groupe de Sarah Ennis et Eleanor Seaby (Univ. of Southampton, UK et Broad Institute, MIT/Harvard, USA), nous avons identifié des variants de novo hétérozygotes du gène DDX17 chez des patients atteints d'un déficit neuroanatomique, fonctionnel et cognitif, parfois associé à des troubles du spectre de l’autisme. Nos résultats, qui impliquent également l'équipe de Julien Courchet (Labo PGNM, Institut NeuroMyoGene, Lyon) indiquent que DDX17 contribue au développement neuronal in vitro et in vivo, et que sa perte de fonction induit une dérégulation de l'expression de nombreux gènes impliqués dans le développement.
Voir notre article en pré-print
Nos objectifs sont d'étudier plus en détail la fonction du gène DDX17, en caractérisant notamment ses gènes et exons cibles pour comprendre sa contribution au développement neuronal, et d'étudier l'impact des mutations du gène DDX17 sur cette fonction.
Etude des liens fonctionnels entre l'oncogène MYCN et les hélicases DDX17 et DDX5 dans le neuroblastome
 Valentine CLERC (doctorante ENS-Lyon)
Valentine CLERC (doctorante ENS-Lyon)
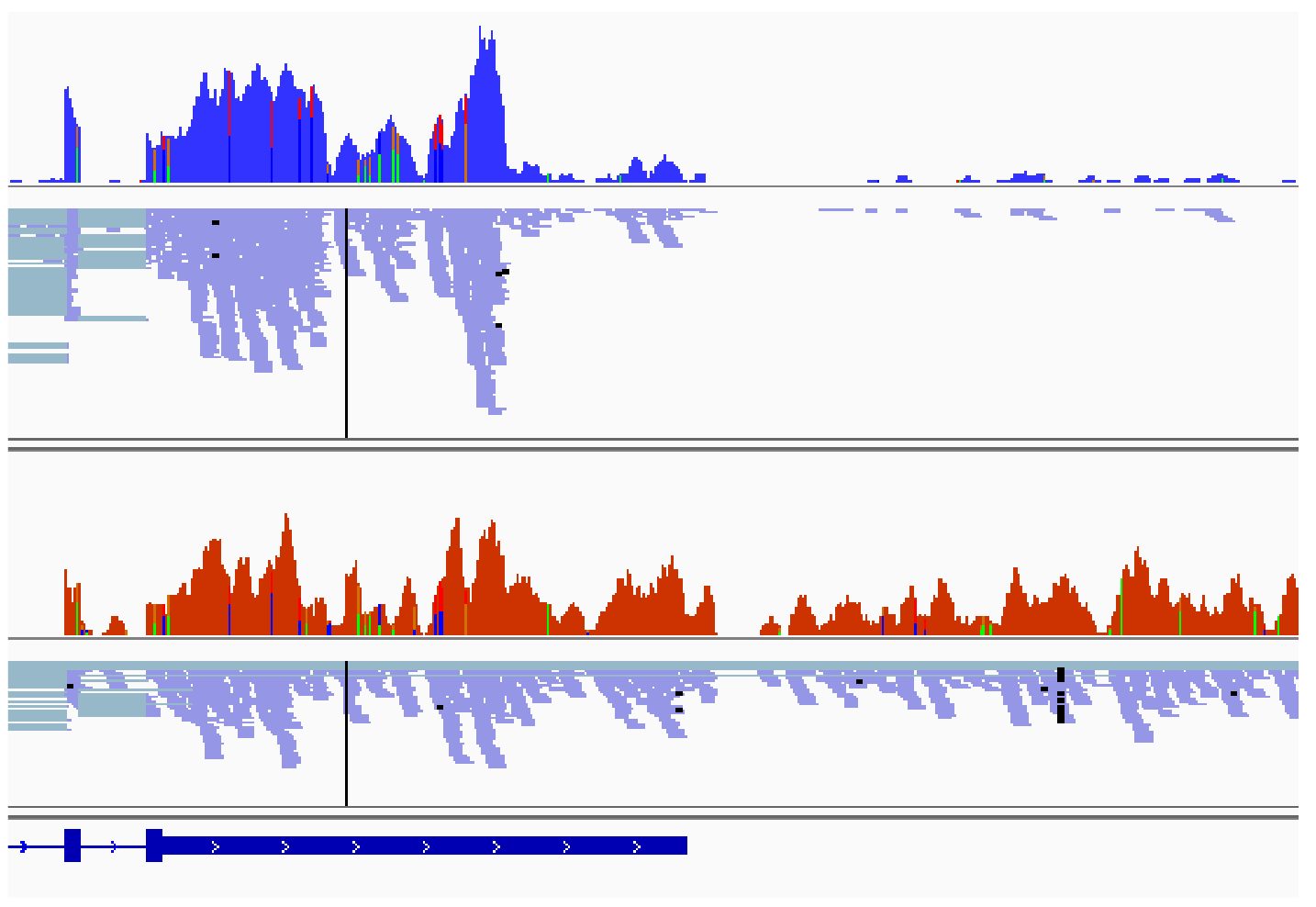
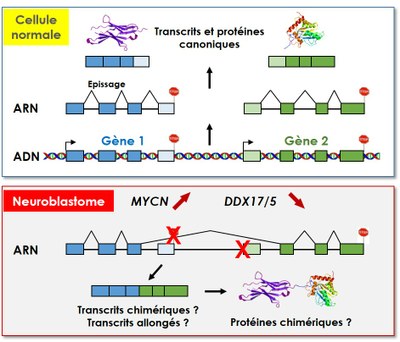
Mon projet de thèse s'intéresse à la régulation de la maturation en 3' des transcrits par DDX17 et DDX5 dans le contexte du neuroblastome. Nous explorons l'hypothèse que l’expression réduite de DDX17 et DDX5 dans les neuroblastomes les plus agressifs soit associée à un contrôle transcriptionnel moins strict et à une augmentation de la production de transcrits aberrants, par exemple des transcrits chimériques, formés par épissage entre des transcrits issus de deux gènes adjacents orientés en tandem. Par ailleurs, j'étudie les liens fonctionnels entre l'expression des hélicases DDX17 et DDX5 et l'oncogène MYCN dans les neuroblastomes.
En collaboration avec l'équipe de Sarah Ennis (Université de Southampton, UK), j'étudie aussi l'impact fonctionnel de mutations récemment identifiées dans le gène DDX17, associées à un désordre neuro-développemental.
Financement : Fondation pour la Recherche Médicale
Bioinformatic analyses
 Hélène Polvèche (Ingénieur d'études en Bio-informatique; IStem – CECS)
Hélène Polvèche (Ingénieur d'études en Bio-informatique; IStem – CECS)
Créée en 2014 au sein de l’Institut des cellules Souches pour le Traitement et l’Étude des Maladies monogéniques (I-Stem) à Evry, la plateforme NGS & analyses génomiques (https://www.istem.eu/plateformes-technologiques/analysesgenomiques/) est une structure collaborative ouverte aux équipes académiques et a été récemment labellisée Genopole.
Depuis 2015, la plateforme est en collaboration avec l’équipe ReGarDS afin de pérenniser la veille bio-informatique en constante évolution et d’explorer de façon poussée des données issues de séquençage. Ce travail collaboratif a notamment permis l’étude avec l’équipe de C. Martinat, de l’impact de l’épissage alternatif dans la dystrophie myotonique de type 1 (DM1) et d’identifier de nouvelles cibles thérapeutiques (Laustriat et al, Mol Ther Nucleic Acids. 2016, Maury et al, iScience. 2019).
En parallèle de ces projets, la plateforme développe de nouveaux outils de séquençage ou d’analyses bio-informatiques. Par exemple, nous avons mis en place une base de données d’expression génique permettant d’étudier l’expression d’un ou de plusieurs gènes dans différents modèles cellulaires dérivés de cellules souches pluripotentes humaines (SISTEMA) ou plus récemment, une base de données et une ressource web permettant de prédire les effets sur l’épissage alternatif de plus d’une cinquantaine de facteurs d’épissage (SplicingLore, lien vers l'article).
* Missions :
- Traitements & Analyses de données (Transcriptomique) issues de RNA-seq / Ampli-seq / Quant-seq (3'-seq) / miRNA-seq (Illumina / Ion Proton)
- Mise en place de Pipelines d'analyses bio-informatiques
- Création d'outils via interfaces web
* Langages de programmation :
Bash, Perl, Python, R, HTML, SQL, PHP, Javascript, jQuery
* Supports / OS :
HPCC Grid Engine (SGE), Linux Debian - Ubuntu – Windows
William Desaintjean (Ingénieur d'études)
William a été recruté dans le cadre d'un projet ANR (ASTROMYOD) coordonné par Mario Gomes-Pereira (Institut de Myologie, Paris), pour conduire des analyses visant à mieux comprendre les altérations du transcriptome dans le système nerveux central dans un modèle murin de la Dystrophie Myotonique de type 1 (DM1). Sa mission principale est de mettre en place et de développer un pipeline d'analyse de données de séquençage pleine longueur (Nanopore), qui permettra notamment d'étudier les variations d'épissage et de maturation en 3' (polyadénylation/terminaison) dans nos modèles d'étude.
* Langages de programmation :
Bash, Python, R, C++
Références
1. Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert MP, Neil H, Zonta E, Mortada H, Gratadou L, Deygas M, Chakrama FZ, Samaan S, Desmet FO, Tranchevent LC, Dutertre M, Rimokh R, Bourgeois CF*, Auboeuf D* (2014). RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation.Cell Rep.7:1900-13 (*corresponding authors). PDF here
2. Bourgeois CF, Mortreux F, Auboeuf D (2016).The multiple functions of RNA helicases as drivers and switchers of the genetic information flow. Nat. Rev. Mol. Cell. Biol.17:426-38.
3. Bourgeois CF, Auboeuf D (2017). The RNA helicase DDX5 is a reprogramming roadblock. Stem Cell Investig.4:79. PDF here
4. Lambert MP, Terrone S, Giraud G, Benoit-Pilven C, Cluet D, Combaret V, Mortreux F, Auboeuf D, Bourgeois CF (2018). The RNA helicase DDX17 controls the transcriptional activity of REST and the expression of proneural microRNAs in neuronal differentiation. Nucleic Acids Res.46:686–7700. PDF here
5. Giraud G, Terrone S., Bourgeois CF (2018). Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep.51: 613-622. PDF here
6. Terrone S, Valat J, Fontrodona N, Giraud G, Claude J, Combe E, Lapendry A, Polvèche H, Ameur LB, Duvermy A, Modolo L, Bernard P, Mortreux F, Auboeuf D, Bourgeois CF (2022). RNA helicase-dependent gene looping impacts messenger RNA processing. Nucleic Acids Res, 50(16):9226-46. PDF here
7. Polvèche H, Valat J, Fontrodona N, Lapendry A, Clerc V, Janczarski S, Mortreux F, Auboeuf D, Bourgeois CF (2023). SplicingLore: a web resource for studying the regulation of cassette exons by human splicing factors. Database 2023 Dec 21. PDF here
Personnel impliqué
Valentine CLERC (doctorat, depuis 2020)
Xavier GRAND (IR bio-informatique, contrat UCBL en co-supervision avec Barbara Testoni, CRCL)
Hélène POLVECHE (IE bio-informatique, CDI I-Stem)
William DESAINTJEAN (IE bio-informatique)
Principales collaborations
Thomas SEXTON (IGBMC, Illkirch/Strasbourg, France): Role of 3D genome organization in RNA processing
Barbara TESTONI (CRCL, Lyon, France): DDX5/DDX17 and infection by Hepatitis B Virus
Geneviève GOURDON et Mario GOMES-PEREIRA (Institut de Myologie, Paris, France): RNA processing alterations in Myotonic Dystrophy
Julien COURCHET (INMG, Lyon, France): RNA processing regulation in the mouse nervous system
Sarah ENNIS (University of Southampton, UK): Functional analysis of de novo DDX17 mutations
Financements récents
2016-2020 ANR CHROTOPAS (ANR16-CE12-0009-01)
2018-2019 Ligue contre le Cancer (Comité de la Loire)
2018-2019 Fondation ARC
2019-2020 ANRS (coordinateur: B. Testoni)
2020-2021 Ligue contre le Cancer (Comité du Rhône)
2020-2021 Association Hubert Gouin, "Enfance et Cancer"
2020-2024 Labellisation Ligue contre le Cancer
2023-2025 ANR ASTROMYOD (coordinateur: M. Gomes-Pereira)
2024-2025 Association Hubert Gouin "Enfance et Cancer"
Principaux anciens membres
Jessica VALAT, PhD (doctorat, 2017-2022) --> Police Technique et Scientifique
Sophie TERRONE, PhD (doctorat, 2015-2019) --> Post-doc
Guillaume GIRAUD, PhD (post-doc, 2017-2019) --> Post-doc
Marie-Pierre LAMBERT, PhD (post-doc, 2013-2016) --> Genomics Consulting
Micaela POLAY-ESPINOZA, PhD (doctorat, 2011-2014)
