Control of gene expression by RNA helicases DDX5 and DDX17 - Cyril Bourgeois
Control of gene expression by RNA helicases DDX5 and DDX17
 Project leader : Cyril BOURGEOIS (cyril.bourgeois@inserm.fr)
Project leader : Cyril BOURGEOIS (cyril.bourgeois@inserm.fr)
ResearcherID E-6337-2010
ORCID 0000-0002-0756-5501
Loop
LinkedIn
Twitter @C_Bourgeois_ENS
DDX17 and DDX5 are two highly related DEAD box ATPases that play various molecular functions contributing to the control of gene expression. They are directly involved in several RNA processing mechanisms (pre-mRNA splicing, processing of microRNA precursors) and our work has highlighted their important role in alternative splicing regulation (1,2,4). DDX17 and DDX5 also control directly the binding and the activity of several transcription factors on chromatin, through unclear mechanisms (4,5). Thus, the multi-layered activity of those proteins allows a dynamic orchestration of gene expression that is crucial for some cell differentiation processes (1-4). For example, we have shown the essential role of DDX17 and DDX5 during myogenic differentiation or during epithelial-to-mesenchymal transition (EMT) (1), a cellular transformation occuring during embryogenesis but also during metastasis. More recently we have also uncovered an important function of DDX17 in the early stages of neuroblastoma cell differentiation (4).
We are interested in the DDX17/DDX5-involving chromatin mechanisms that control the overall transcriptional process (RNA Polymerase II-mediated transcription itself, but also co-transcriptional steps such as splicing and polyadenylation). We have recently shown that DDX17/DDX5 are involved in the spatial folding of genes, and that chromatin looping has an impact on RNA processing in human cells (6). Beside traditional techniques of molecular and cellular biology (cell transfection, analysis of RNA and protein expression…), we are using more exploratory approaches allowing to specifically modify the genome (CRISPR-Cas9) or to analyse the 3D structure of genes (3C/4C). Our research exploits various NGS datasets (RNA-seq, ChIP-seq…) through computational approaches that allow to look into these mechanisms from a wider perspective.
In the continuity of our previous work, our objectives are to better understand at the molecular level how DDX5 and DDX17 regulate the expression of genes and the maturation of pre-messenger RNA. The projects of my group are articulated around 2 main axes linked to two different types of pathologies having in common an alteration of the expression or function of DDX17: a cancer (neuroblastoma) and a genetic disease (a neurodevelopmental syndrome associated with mutations of the DDX17 gene).
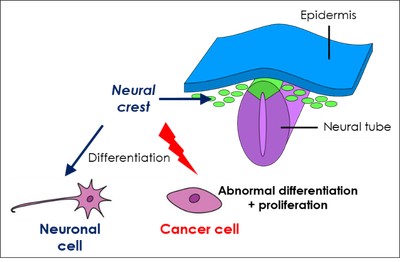
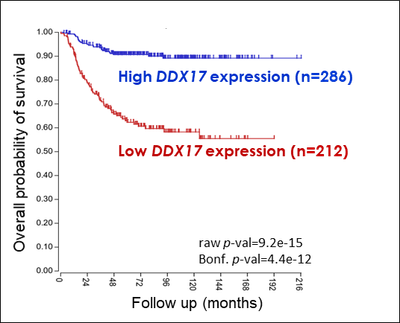
1. Neuroblastoma is a pediatric cancer that is often uncurable in its most serious forms. An essential aspect of the development of neuroblastomas to more or less aggressive forms is the capacity of cancer cells, originating from the peripheral nervous system, to proliferate in an aberrant manner or to differentiate into benign neuronal cells, a process very difficult to control therapeutically. DDX17 plays a major role in the differentiation of neuroblastoma cells (4), and its expression has a good prognostic value for this cancer. These results suggest that a reduced expression of these factors, which leads in cellulo to deep transcriptomic (non genomic) alterations, may contribute to the development of the most aggressive neuroblastomas, an hypothesis we are investigating.
2. Impact of DDX17 gene mutations in a neurodevelopmental syndrome. In collaboration with the group of Sarah Ennis and Eleanor Seaby (Univ. of Southampton, UK and Broad Institute, MIT/Harvard, USA), we have identified de novo heterozygous variants of the DDX17 gene in patients with neuroanatomical, functional and cognitive deficits, sometimes associated with autism spectrum disorders. Our as yet unpublished results, which also involve Julien Courchet's team (PGNM Lab, NeuroMyoGene Institute, Lyon), indicate that DDX17 contributes to neuronal development and that its loss of function induces a deregulation of the expression of numerous genes involved in development.
Link to our pre-print article
Our objectives are to study in more detail the function of the DDX17 protein, in particular by characterising its target genes and exons in order to understand its contribution to neuronal development, and to study the impact of DDX17 gene mutations on this function.
Functional relationship between MYCN oncogene and helicases DDX17 and DDX5 in neuroblastoma
 Valentine CLERC (PhD student, ENS-Lyon)
Valentine CLERC (PhD student, ENS-Lyon)
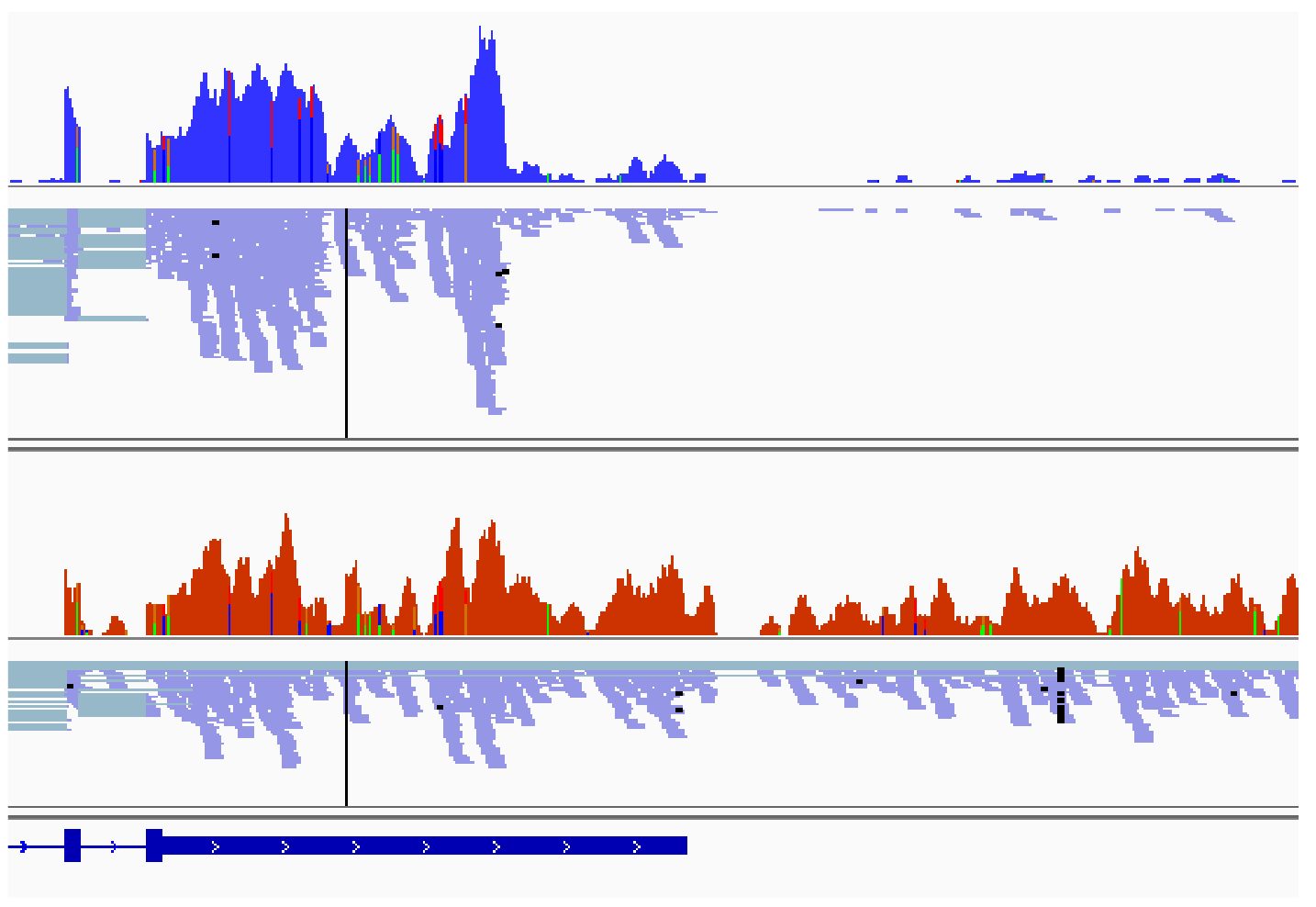
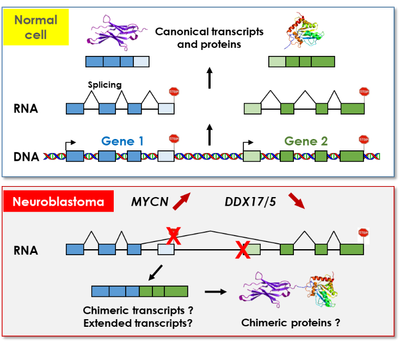
My PhD project is focussed on the regulation of 3' end processing of mRNA by DDX17 and DDX5 in the context of neuroblastoma. We are exploring the hypothesis that a reduced expression of DDX17 and DDX5 in agressive neuroblastomas could be associated to an alteration of transcription fidelity and to an increase in the production of aberrant transcripts (like chimeric transcripts that are formed through spliing between exons from adjacent tandem-orientated genes). I am also investigating the functional relationship between the expression of DDX17 and DDX6 and the oncogene MYCN in neuroblastoma cells.
In collaboration with the team of Sarah Ennis (University of Southampton, UK), I am also investigating the functional impact of mutations recently identified in the DDX17 gene, associated to a neuro-developmental disorder.
Funding : PhD fellowship from Fondation pour la Recherche Médicale
Bioinformatic analyses
 Hélène Polvèche (Computational biologist; IStem – CECS)
Hélène Polvèche (Computational biologist; IStem – CECS)
Created in 2014 within the Institute of Stem Cells for the Treatment and Study of Monogenic Diseases (I-Stem) in Evry, the NGS & genomic analysis platform (https://www.istem.eu/plateformes-technologiques/analysesgenomiques/) is a collaborative structure open to academic teams and was recently awarded Genopole accreditation. Since its creation, the platform has sequenced various transcriptomes and helped teams to analyse these projects (Darville et al, EBioMedicine 2016; Cambon et al, Molecular therapy. Methods & clinical development 2017; Galvan et al, Brain 2018; Hoch et al, Sci Rep. 2019).
Since 2015, the platform has been collaborating with the ReGarDS team in order to perpetuate the constantly evolving bioinformatics watch and to explore sequencing data in depth. In particular, this collaborative work has made it possible to study, with C. Martinat's team, the impact of alternative splicing in myotonic dystrophy type 1 (DM1) and to identify new therapeutic targets (Laustriat et al, Mol Ther Nucleic Acids. 2016, Maury et al, iScience. 2019).
Alongside these projects, the platform is developing new sequencing and bioinformatics analysis tools. For example, we have set up a gene expression database to study the expression of one or more genes in different cell models derived from human pluripotent stem cells (SISTEMA) and, more recently, a database and web resource to predict the effects on alternative splicing of more than fifty splicing factors (SplicingLore, link to article).
* Missions :
- Processing & Analysis of data (Transcriptomics) from RNA-seq / Ampli-seq / Quant-seq (3'-seq) / miRNA-seq (Illumina / Ion Proton)
- Setting up bioinformatics analysis pipelines
- Creation of tools via web interfaces
* Programming languages :
Bash, Perl, Python, R, HTML, SQL, PHP, Javascript, jQuery
* Supports / OS :
HPCC Grid Engine (SGE), Linux Debian - Ubuntu – Windows
William Desaintjean (Computational biologist)
William has been recruited as part of an ANR project (ASTROMYOD) coordinated by Mario Gomes-Pereira (Institut de Myologie, Paris), to conduct analyses aimed at gaining a better understanding of transcriptome alterations in the central nervous system in a mouse model of Myotonic Dystrophy type 1 (DM1). His main task is to set up and develop a pipeline for analysing full-length sequencing data (Nanopore), which will make it possible to study variations in splicing and 3' maturation (polyadenylation/termination) in our models.
* Programming languages :
Bash, Python, R, C++
References
1. Dardenne E, Polay Espinoza M, Fattet L, Germann S, Lambert MP, Neil H, Zonta E, Mortada H, Gratadou L, Deygas M, Chakrama FZ, Samaan S, Desmet FO, Tranchevent LC, Dutertre M, Rimokh R, Bourgeois CF*, Auboeuf D* (2014). RNA helicases DDX5 and DDX17 dynamically orchestrate transcription, miRNA, and splicing programs in cell differentiation.Cell Rep.7:1900-13 (*corresponding authors). PDF here
2. Bourgeois CF, Mortreux F, Auboeuf D (2016).The multiple functions of RNA helicases as drivers and switchers of the genetic information flow. Nat. Rev. Mol. Cell. Biol.17:426-38.
3. Bourgeois CF, Auboeuf D (2017). The RNA helicase DDX5 is a reprogramming roadblock. Stem Cell Investig.4:79. PDF here
4. Lambert MP, Terrone S, Giraud G, Benoit-Pilven C, Cluet D, Combaret V, Mortreux F, Auboeuf D, Bourgeois CF (2018). The RNA helicase DDX17 controls the transcriptional activity of REST and the expression of proneural microRNAs in neuronal differentiation. Nucleic Acids Res.46:686–7700. PDF here
5. Giraud G, Terrone S., Bourgeois CF (2018). Functions of DEAD box RNA helicases DDX5 and DDX17 in chromatin organization and transcriptional regulation. BMB Rep.51: 613-622. PDF here
6. Terrone S, Valat J, Fontrodona N, Giraud G, Claude J, Combe E, Lapendry A, Polvèche H, Ameur LB, Duvermy A, Modolo L, Bernard P, Mortreux F, Auboeuf D, Bourgeois CF (2022). RNA helicase-dependent gene looping impacts messenger RNA processing. Nucleic Acids Res, 50(16):9226-46. PDF here
7. Polvèche H, Valat J, Fontrodona N, Lapendry A, Clerc V, Janczarski S, Mortreux F, Auboeuf D, Bourgeois CF (2023). SplicingLore: a web resource for studying the regulation of cassette exons by human splicing factors. Database 2023 Dec 21. PDF here
People involved
Valentine CLERC (PhD student, arrived in 2020)
Xavier GRAND, PhD (computational biologist, arrived in 2021, contract UCBL in co-supervision with Barbara Testoni, CRCL)
Helène POLVECHE (computational biologist, long-term contract with I-Stem institute)
William DESAINTJEAN (computational biologist)
collaborations
Thomas SEXTON (IGBMC, Illkirch/Strasbourg, France): Role of 3D genome organization in RNA processing
Barbara TESTONI (CRCL, Lyon, France): DDX5/DDX17 and infection by Hepatitis B Virus
Geneviève GOURDON et Mario GOMES-PEREIRA (Institut de Myologie, Paris, France): RNA processing alterations in Myotonic Dystrophy
Julien COURCHET (INMG, Lyon, France): RNA processing regulation in the mouse nervous system
Sarah ENNIS (University of Southampton, UK): Functional analysis of de novo DDX17 mutations
recent funding
2016-2020 ANR CHROTOPAS (ANR16 CE12-0009-01)
2018-2019 Ligue contre le Cancer (Comité de la Loire)
2018-2019 Fondation ARC
2019-2020 ANRS
2020-2021 Ligue contre le Cancer (Comité du Rhône)
2020-2021 Association Hubert Gouin, "Enfance et Cancer"
2020-2024 Labellisation Ligue contre le Cancer
2023-2025 ANR ASTROMYOD (coordinateur: M. Gomes-Pereira)
2024-2025 Association Hubert Gouin, "Enfance et Cancer"
Main past members
Jessica VALAT, PhD (PhD student, 2017-2022) --> Technical and Scientific Police
Sophie TERRONE, PhD (PhD student, 2015-2019) --> Post-doc
Guillaume GIRAUD, PhD (post-doc, 2017-2019) --> Post-doc
Marie-Pierre LAMBERT, PhD (post-doc, 2013-2016) --> Genomics Consulting
Micaela POLAY-ESPINOZA, PhD (PhD student, 2011-2014)
