Molecular mechanism of homology search
Homologous recombination (HR) is an elaborate DNA break repair pathway that uniquely exploits an intact homologous double-strand DNA (dsDNA) molecule as a template for repair. The use of this intact “donor” can entail a genome- and nucleus-wide search for homology by the broken molecule, in the form of a nucleoprotein filament. In eukaryotes, this filament is composed of a helical filament of Rad51 and associated proteins that harnesses a ssDNA sequence up to several kilobases-long to sample in parallel the dsDNA molecules colliding with it in space (Figure 1). Despite decades of study, the mechanism by which a single broken molecule achieves accurate and efficient identification of a single homologous dsDNA located anywhere in the heterologous haystack of the genome remains a main conundrum of the DNA repair field.

Figure 1: Inter-segmental contact sampling of dsDNA by the Rad51-ssDNA filament.
The nucleoprotein factors implicated and their quantitative contribution to homology sampling remains ill-defined in vivo. By using a a combination of homemade quantitative in vivo methodologies and in silico modelling we try achieving a quantitative understanding of the search process in cells.
This methodology and a highly efficient site-specific DNA break induction system in S. cerevisiae cells enable us to follow the kinetics of formation of various HR intermediates and steps at the population level (Figure 2), in various sequence and mutant contexts. These experimental data can be compared with the results of in silico simulations to understand the quantitative contributions of the factors under scrutiny.
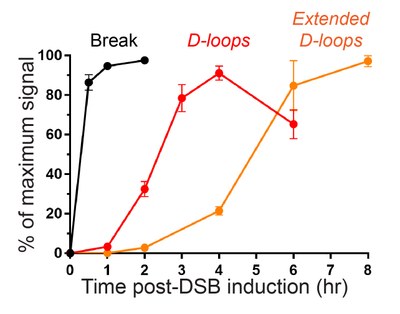
Figure 2: Example of the kinetics of inter-chromosomal D-loop formation and extension at a 2 kb-long region of homology in wild-type yeast cells.
