Automated Analyis of quantitative Yeast Two-Hybrid
This program permits to easily generate in cellulo affinity ladders from quantitative Yeast Two-Hybrid experiments.


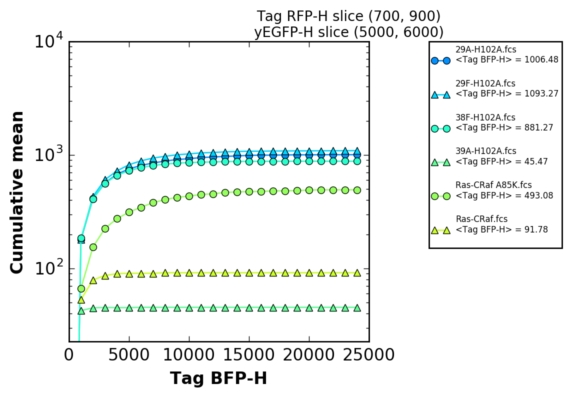
This program has been developped to permits rapid generation of in-vivo affinity ladder from quantitative Yeast Two-Hybrid experiments [ref].

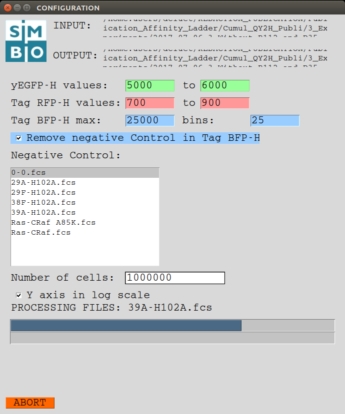
REQUIREMENTS:
This program is optimized for Python 2.7 with the following libraries:
- datetime: To generate unique Analysis ID and file name.
- FlowCytometryTools: To open `.fcs` files and manipulate flowcytometry data [Website].
- glob: To identify the `.fcs` files in the Input folder.
- matplotlib: To generate the curves.
- numpy: To generate and manipulate arrays.
- os: To handle paths of the raw data and generated files.
- Pillow / PIL: To display images within the GUI of the program.
- sys: To permit manual abortion of the program.
- Tkinter: To generate the GUI of the program.
Download:
Test Files:
- Negative control (Download)
- Covalent normalization control (Download)
- Barstar Y29F - Barnase H102A, Kd=117 pM (Download)
- Barstar Y29A - Barnase H102A, Kd=420 pM (Download)
- Barstar W38F - Barnase H102A, Kd=4 000 pM (Download)
- Ras G12V C186A - C-Raf RBD A85K, Kd=11 000 pM (Download)
- Ras G12V C186A - C-Raf RBD, Kd=122 000 pM (Download)
-
Barstar D39A - Barnase H102A, Kd=420 000 pM (Download)
