RESOURCES :
How plant cells positioned their division planes during morphogenesis?
During the life cycle of any multicellular organism, cell division contributes to the generation of specialized cells, which are necessary to form tissues and carry out particular functions. Therefore, the mechanisms of cell division need to be tightly regulated, as malfunctions in their control can lead to tumour formation or developmental defects. This is particularly true in land plants, where cells cannot migrate and therefore cytokinesis is key for morphogenesis. In plant, cell division is executed in radically different manners than animals, with the appearance of new structures (cell plate, cytokinetic phragmoplast and the preprophase band (PPB) of microtubules) and the disappearance of ancestral mechanisms (cleavage, centrosomes).
A crucial feature of cell division is how cells position their division plane. It has even more significance for walled organisms, because their cells are embedded and cannot relocate. The memory of the cell division plane is maintained throughout plant cell division by a landmark at the equatorial periphery of the cell. Recent evidence shows that membrane lipids such as phosphoinositides (PIPs) are important for cell division orientation in plants. Phosphoinositides are ubiquitous regulators of signal transduction events in eukaryotic cells. They are produced by mono-, bis- and trisphosphorylation of the inositol head group of phosphatidylinositol. Due to their charged nature, phosphoinositides may create localized docking sites for proteins—for example, cytoskeletal and trafficking regulators—thereby defining specific membrane domains.
We are currently investigating the interplay between the phosphoinositides and the cytoskeleton during plant cell division using Arabidopsis and BY2-cells as models.
We want now to address:
-
•What is the crosstalk between the phosphoinositides at the plasma membrane and the cytoskeleton during mitosis ?
-
•How the phosphoinositides-patterning, together with the cytoskeleton, are organized in dividing root cells ?
-
•What is the impact of local perturbation of the phosphoinositides-patterning on cell division orientation ?
-
•What are the upstream components responsible for the distribution of the phosphoinositides during cytokinesis ?
Results gained from the ongoing research will provide a better understanding of the signaling mechanisms coupling PIP-metabolism and cell division, the latest being fundamental in regulating organismal growth.

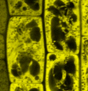
Our research is based on cell and developmental biology approaches, such as live confocal imaging, expression pattern analysis, mutant phenotype description, genetics as well as protein biochemistry

VINCENT BAYLE (Engineer)
FREDERIQUE ROZIER (ENGINEER)
AURELIE FANGAIN (ENGINEER)
ALEXIS LEBECQ (PHD STUDENT)
KEY PUBLICATIONS
Cell Signaling lab
RDP - ENS Lyon - 46 Allee d’italie
69364 Lyon cedex 07 - France
Web Site : Yvon Jaillais


- Plant Biology - Cell Biology - Plant development - Cell Signaling - Signal transduction - Receptor Kinase - Plant adaptation - Endocytosis - Self Incompatibility - Plant Reproduction - Brassinosteroid - Plant hormones - hormone crosstalk - Intracellular trafficking - Live confocal imaging - BRI1- BKI1- SRK - SCR - Retromer - SNX - Sorting Nexin - Vacuolar Protein Sorting - VPS - RDP, ENS Lyon, Ecole Normale Superieure - Laboratoire de Reproduction et development des plantes - France -



OUR RESEARCH IS FUNDED BY:
Alexis Lebecq, Aurélie Fangain, Alice Boussaroque, Marie-Cécile Caillaud. Dynamic Apical-Basal Enrichment of the F-Actin during Cytokinesis in Arabidopsis Cells Embedded in their Tissues. Quantitative Plant Biology. in press (2021)
on bioRxiv 2021.07.07.451432; doi: https://doi.org/10.1101/2021.07.07.451432
Mehdi Doumane, Léia Colin, Alexis Lebecq,
Aurélie Fangain, FD Stevens, Joseph Bareille,
Olivier Hamant, Youssef Belkhadir, T Munnik,
Yvon Jaillais #, Marie-Cécile Caillaud #.
Inducible depletion of PI(4,5)P2 by the
synthetic iDePP system in Arabidopsis.
Nature Plants 7 587-597 (2021)
on bioRxiv 2020.05.13.091470;
doi: https://doi.org/10.1101/2020.05.13.091470
Platre, M.P., Noack, L.C., Doumane, M., Bayle, V.,
Simon, M.L.A., Maneta-Peyret, L., Fouillen, L., Stanislas, T.,
Armengot, L., Pejchar, P., Caillaud, M.C., Potocky, M.,
Copic, A., Moreau, P., and Jaillais, Y.2018.
A combinatorial lipid code shapes the electrostatic
landscape of plant endomembranes. Dev Cell
21;45(5):645-480.e11
open access version on biorxiv:
A combinatorial lipid code shapes the electrostatic
landscape of plant endomembranes. bioRxiv, p.278135.
Doumane M, Lionnet C, Bayle V, Jaillais Y, Caillaud MC#. Automated Tracking of Root for Confocal Time-lapse Imaging of Cellular Processes. Bio Protoc. 2017 Apr 20;7(8). pii: e2245. doi: 10.21769/BioProtoc.2245. PMID: 28459086
Simon ML*, Platre MP*, Marquès-Bueno MM, Armengot L, Stanislas T, Bayle V, Caillaud MC#, Jaillais Y#. A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nature Plants. 2016 Jun 20;2:16089. doi: 10.1038/nplants.2016.89. PMID: 27322096
Caillaud MC*, Wirthmueller L*, Sklenar J, Findlay K, Piquerez SJ, Jones AM, Robatzek S, Jones JD, Faulkner C. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 2014 Nov 13;10(10):e1004496.
Caillaud MC, Asai S, Rallapalli G, Piquerez S, Fabro G, Jones JD. A downy mildew effector attenuates salicylic Acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 2013 Dec;11(12):e1001732
Caillaud MC, Piquerez SJ, Fabro G, Steinbrenner J, Ishaque N, Beynon J, Jones JD. Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility.Plant Journal. 2012 Jan;69(2):252-65.
Caillaud MC, Piquerez SJ, Jones JD. 2012. Characterization of the membrane-associated HaRxL17 Hpa effector candidate. Plant Signal Behav 7: 145-9
Fabro G, Steinbrenner J, Coates M, Ishaque N, Baxter L, Studholme DJ, Korner E, Allen RL, Piquerez SJ, Rougon-Cardoso A, Greenshields D, Lei R, Badel JL, Caillaud MC, Sohn KH, Van den Ackerveken G, Parker JE, Beynon J, Jones JD. 2011. Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathog 7: e1002348
Caillaud MC, Paganelli L, Lecomte P, Deslandes L, Quentin M, Pecrix Y, Le Bris M, Marfaing N, Abad P, Favery B. 2009. Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS One 4: e6757
Caillaud MC, Abad P, Favery B. 2008. Cytoskeleton reorganization, a key process in root-knot nematode-induced giant cell ontogenesis. Plant Signal Behav 3: 816-8
Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier TR, Markov GV, McVeigh P, Pesole G, Poulain J, Robinson-Rechavi M, Sallet E, Segurens B, Steinbach D, Tytgat T, Ugarte E, van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. 2008. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol 26: 909-15
Caillaud MC, Lecomte P, Jammes F, Quentin M, Pagnotta S, Andrio E,
de Almeida Engler J, Marfaing N, Gounon P, Abad P, Favery B. 2008.
MAP65-3 microtubule-associated protein is essential for nematode-induced
giant cell ontogenesis in Arabidopsis. Plant Cell 20: 423-37
Caillaud MC, Dubreuil G, Quentin M, Perfus-Barbeoch L, Lecomte P,
de Almeida Engler J, Abad P, Rosso MN, Favery B. 2008.
Root-knot nematodes manipulate plant cell functions during a compatible
interaction. J Plant Physiol 165: 104-13
ANR “INTERPLAY” ANR-16-CE13-0021
ANR “Divcon” 2022-2026
in collaboration with Emmanuel Bayer