RESOURCES :
How individual cells communicate with each other and with the environment to shape plant architecture?
Plants are sessile organisms and therefore must constantly adapt their growth and architecture to an ever-changing environment. It is the delicate balance between developmental and environmental signals that shapes the architecture of the plants. Individual cells constantly integrate those cues into cellular output, that gives rise to new organs. Cells are equipped with receptor molecules that allow communication with other cells in the organism and detection of changes in their surrounding environment, such as changes in light quality, nutrient availability, attack by pathogens...
The plasma membrane plays a critical role in regulating exchanges between the intra- and extracellular milieu and in controlling cell-to-cell communication. The aim of our group is to understand what makes the plasma membrane unique with regard to other membrane compartments and as such competent for signaling. We want to understand how signaling domains are being defined at the plant plasma membrane, both at the level of the cell (e.g., polar domains) but also at the micrometer scale (e.g., membrane microdomains). We also plan to address how these signaling domains evolve during the course of signaling and what are their functional roles during cell-cell communication.
Anionic phospholipids (e.g. phosphoinositides, PIPs) are minor lipids in membranes but they have a huge impact on cell signaling and organelle identity. They not only recruit proteins to specific cellular compartments, but they also deeply impact membrane biophysics properties. Furthermore, their production is tightly and dynamically regulated during development and interaction with the environment.
We want to address :
- What are the anionic lipids involved in plasma membrane organization and how?
-
-What are the functions of anionic lipids and their interacting proteins in hormone signaling?
(e.g. brassinosteroid and auxin signaling)
-
-How do anionic phospholipids contribute to endomembrane compartment identity and orchestrate intracellular trafficking?
- How is anionic lipids homeostasis regulated by changes in the environment and contribute to plant adaptation?

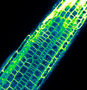
Our research is based on cell and developmental biology approaches, such as live confocal imaging, expression pattern analysis, mutant phenotype description, genetics as well as protein biochemistry

OUR RESEARCH IS FUNDED BY:
CHRISTINE MIEGE (LECTURER)
VINCENT BAYLE (Engineer)
FREDERIQUE ROZIER (ENGINEER)
CHLOE BEZIAT (POST-doc)
LISE NOACK (PHD STUDENT)
GWENNOGAN DUBOIS (PHD St)
ANR “STAYING TIGHT”
in collaboration with Emmanuelle Bayer
KEY PUBLICATIONS
Cell Signaling lab
RDP - ENS Lyon - 46 Allee d’italie
69364 Lyon cedex 07 - France
Web Site : Yvon Jaillais


- Plant Biology - Cell Biology - Plant development - Cell Signaling - Signal transduction - Receptor Kinase - Plant adaptation - Endocytosis - Self Incompatibility - Plant Reproduction - Brassinosteroid - Plant hormones - hormone crosstalk - Intracellular trafficking - Live confocal imaging - BRI1- BKI1- SRK - SCR - Retromer - SNX - Sorting Nexin - Vacuolar Protein Sorting - VPS - RDP, ENS Lyon, Ecole Normale Superieure - Laboratoire de Reproduction et development des plantes - France -














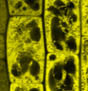
ANR “caLIPSO”
in collaboration with Yohann Boutte
CIG “RLK-negreg” (2012-2016)
PCIG10-GA-2011-303601
ERC starting grant “APPL”
336360-APPL (2014-2019)
ERA CAPS “SICOPID”
in collaboration with:
Michael Hothorn, University of Geneva, Switzerland
Cyril Zipfel, Univeristy of Zurich, Switzerland
Zach Nimchuk, Unibersity of North Carolina, Chapel Hill, USA
Thorsten Nurnberger, ZMBP, Tubingen, Germany

PAST FUNDING:
ANR JCJC “RIPL-MAKR” 2013
ERC consolidator grant “LIPIDEV”
(2021-2026)