RNA polymerases and R-loops in chromosome assembly and integrity-V. Vanoosthuyse
Interplays between transcription and condensin
Transcription can constitute a threat to genome stability by competing with DNA-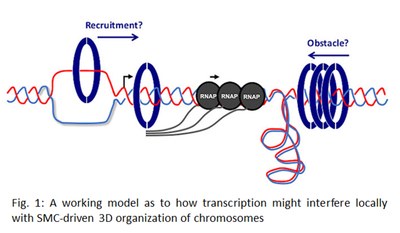 bound machineries or interfering with other types of DNA transactions. For example, growing evidence strongly suggests that transcription can interfere locally with genome duplication by DNA polymerases or with the SMC-driven 3D organization of chromosomes (Fig. 1). The features of transcription that are directly responsible for these genome-destabilizing threats are still under investigation and are the primary focus of our research. We use both human cells and fission yeast as model systems for our studies.
bound machineries or interfering with other types of DNA transactions. For example, growing evidence strongly suggests that transcription can interfere locally with genome duplication by DNA polymerases or with the SMC-driven 3D organization of chromosomes (Fig. 1). The features of transcription that are directly responsible for these genome-destabilizing threats are still under investigation and are the primary focus of our research. We use both human cells and fission yeast as model systems for our studies.
In particular, we investigate: how the efficiency of RNA polymerase elongation can influence the distribution of the SMC complex condensin during mitotic chromosome condensation and, how the transcription by-product R-loop can interfere with chromosome integrity.
RNA polymerases as positioning devices for condensin
The highly-conserved SMC complex condensin drives the abrupt and complete reorganization of chromosomes that occurs in early mitosis. In every organism where the distribution of condensin has been determined, it was shown that it accumulates in the vicinity of highly-expressed genes. It is still unclear however whether or not such a localized accumulation of condensin at discrete loci has an impact on the condensin-driven reorganization of chromosomes in mitosis. In addition, the features of transcription driving this accumulation remain under debate. Using genetics and genome editing strategies in fission yeast, we have accumulated compelling evidence that the mechanisms of transcription termination at the 3’ end of genes play a role in determining the local occupancy of condensin, whatever the RNA polymerase involved. More specifically, our experiments indicate that the backtracking of RNA polymerases that occurs naturally at termination sites can drive the accumulation of condensin (Fig. 2). Our data are consistent with the idea that above a certain density threshold, tightly-bound proteins can impede the translocation of condensin on chromatin. It remains to be determined whether this helps or rather dampens the activity of condensin.
Innovative approaches to map and manipulate R-loops in vivo
A by-product of transcription that is of a particular interest to explain the genome-destabilizing properties of transcription are R-loops. R-loops are three-stranded structures that form when the nascent RNA hybridizes with its DNA template, leaving the non-template DNA strand unpaired (Fig. 3). Their formation is likely to be a tell-tale sign of strong local topological constraints in the DNA. Recent evidence suggests that the stabilization of at least a particular subset of R-loops is associated with DNA damage and genome rearrangements. R-loops have also recently been linked to the formation of Common Fragile Sites (CFS) on mitotic chromosomes. However, the molecular characteristics of cytotoxic R-loops are not yet precisely defined.
Understanding the molecular mechanisms by which R-loops drive genome destabilization requires the development of techniques enabling the precise and quantitative genome-wide mapping of R-loops in the different phases of the cell-cycle. In addition, it is essential to develop tools to precisely manipulate R-loop levels whilst limiting the possibility of indirect effects. We are currently developing innovative approaches to do just that using human cells as a model system.
Does the 3D organization of R-loops contribute to their toxicity?
Using Atomic Force Microscopy (AFM), we have shown that, at least in vitro, different R-loops adopt distinct 3D conformations (Fig. 4). We have also shown that the 3D organization of an R-loop is likely to be determined by the ability of its extruded non-template strand to form secondary structures (in red on Fig. 3). Importantly, we have engineered a mutant allele of the mouse Airn gene, which, upon transcription, forms a similar amount of R-loops of comparable sizes to the wild-type gene, but whose 3D organization is profoundly altered and almost undetectable by AFM. This provides us with a perfect tool to address the role played by the 3D organization of R-loops in their toxicity in vivo.
For all enquiries, please contact vincent.vanoosthuyse [at] ens-lyon.fr
There are currently no items in this folder.
