Physics of Biological Systems
We investigate a variety of biological systems at differents scales :
-
DNA protein interactions (Rloops) (C. Faivre-Moskalenko)
Collab. Vincent Vanoosthuyse (LBMC, ENS Lyon)
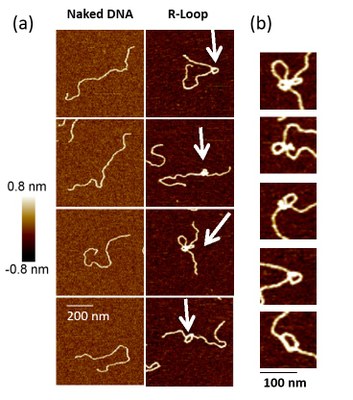
5% of chromosomes are formed by "R-Loops", a DNA/RNA hybrid resulting from transcription, some of which are toxic. Using AFM, we visualize at the single molecule scale R-Loops produced by in vitro transcription. Our results suggest that R-Loops adopt a particular conformation that introduces architectural stress into the DNA.
-
Transport through artificial and natural nanopores (nuclear pore complex) (F.Montel)
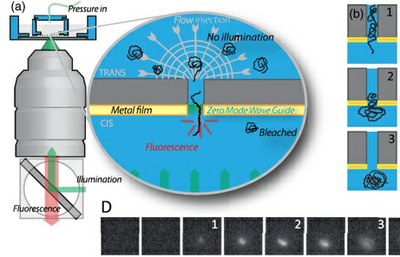
The nuclear pore complex (NPC) is a macromolecular structure which forms the unique gate between the cell nucleus and cytoplasm. The two major characteristics of this biological nanopore are its very high selectivity and directionality. We investigate with minimetic and in vivo approaches the structure and the dynamics of this natural nanopump. We have also develloped single molecules techniques to quantify the translocation of biomolecules in real time in artificial and natural nanopores.
-
Virus mechanics and diassembly (C. Faivre-Moskalenko, M. Castelnovo)
Collab. Anna Salvetti (CRCL, Inserm, Lyon)
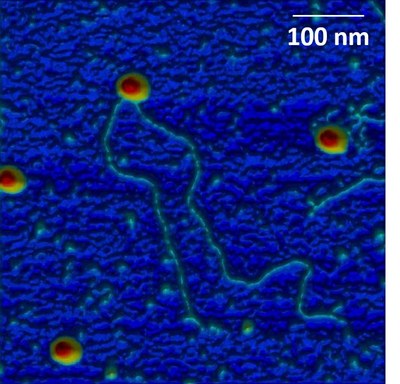
Adeno-Associated Virus (AAV) is a nonpathogenic parvovirus which is currently used as a gene transfer vector in gene therapy applications. Using an original combination of experimental approaches based on AFM and statistical physics modelization, we study viral capsids and AAV particles in particular to define the parameters leading to capsid disassembly and ejection of the viral genome.
-
Bacterial biofilms dynamics (S. Lecuyer)
We are interested in the way swimming bacteria switch to an adhered state, and how adhesion becomes irreversible to initiate a biofilm. We develop innovative experimental approaches to investigate this process, focusing in particular on the role of mechanical signals.
-
Bacterial motility near a surface (C. Place)
For most pathogenic bacteria, flagellar motility is recognized as a virulence factor. Here, we analysed the swimming behaviour of bacteria close to eukaryotic cellular surfaces, using the major opportunistic pathogen Pseudomonas aeruginosa as a model. We delineated three classes of swimming trajectories on both cellular surfaces and glass that could be differentiated by their speeds and local curvatures, resulting from different levels of hydrodynamic interactions with the surface. Thus, bacteria swim near boundaries with diverse patterns and importantly, Type IV pili differentially influence swimming near cellular and abiotic surfaces.
-
Multiscale dynamics of cell nucleus (E. Freyssingeas)
Progress in cellular biology based on fluorescent microscopy techniques, shows that the spatial organization of the nucleus is dynamic. This dynamic is very complex and involves a multitude of phenomena that occur on very different time and size scales. Using an original light scattering experimental device, we investigated the global internal dynamics of the nucleus of a living cell according to the phases of the cell cycle. This dynamic presents two different and independent kinds of relaxation that are well separated in time and specific to the phase of the cell cycle.
-
Chromatin and epigenetics (C. Vaillant, R. Everaers)
Recent advances in genome-wide mapping and imaging techniques have strikingly improved the resolution at which nuclear genome folding can be analyzed and revealed numerous conserved features organizing the one-dimensional chromatin fiber into tridimensional nuclear domains. Understanding the underlying mechanisms and the link to gene regulation requires a crossdisciplinary approach that combines the new high-resolution techniques with computational modeling of chromatin and chromosomes.



