Accueil du site > Thèmes de recherche > Physics to study the living. Project leader : Christophe Place. > Présentation
Présentation
par Webmaster - 2 avril 2012
Our objective is to lead projects at the interface of Physics and Biology by developing original technological approaches. Our main interest, covering different thematic areas, relates to (1) the analysis of the single protein behaviour in interaction with immobilized DNA and, more precisely, on single DNA transcription by T7 RNA polymerase observed by fluorescence microscopy (2) the question of disentangling of long DNA by studying the increase in disentangling velocity in the presence of topoisomérase II using high frequency rheometer, (3) the characterisation of the whole dynamics of living cell nucleus by the dynamic observation of the scattered light by a nucleus (4) the description and the hydrodynamic modeling of the behaviour of motile bacteria in the vicinity of surface using videomicroscopy and Total Internal Reflection Fluorescence Microscopy (TIRFM). The developed techniques are based on soft matter physics and rely on experiments in biochemistry, microbiology and cell biology.
Notre objectif est de mener des projets à l’interface de la Physique et de la Biologie en développant des approches technologiques originales. Nos centres d’intérêts, thématiquement variés, portent sur (1) l’analyse du comportement de protéines uniques en interaction avec de l’ADN immobilisé et notamment sur la transcription d’ADN unique par l’ARN polymérase du phage T7 observée par microscopie de fluorescence, (2) les problèmes de désenchevêtrement d’ADN long tel que l’ADN génomique au cours de la mitose, en rendant compte de l’accélération du processus de désenchevêtrement par l’action de la topoisomérase II par des études de rhéologie haute fréquence et (3) la description et la modélisation hydrodynamique du comportement de bactéries motiles, E. coli, au voisinage de surfaces minérales par vidéomicroscopie et microscopie de fluorescence à réflexion totale interne (TIRF). Les techniques développées sont issues de la physique et s’appuient sur des manipulations en biochimie, microbiologie et biologie cellulaire.
Team Leader ( Chef D’Equipe ) :
- Christophe PLACE - Research Associate First Class
Team Members ( Membres de l’Equipe ) :
- Elodie Chatre - Engineer
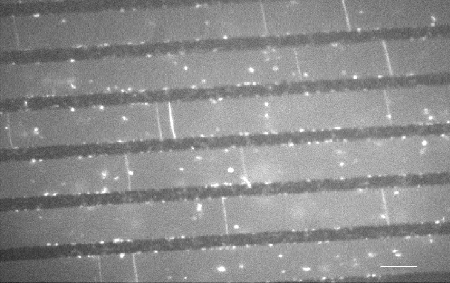
Single T7 DNA are bridged over a micropatterned surface. The black lines are 250 nm above the surface floor (grey). The DNA are visualized by fluorescence microscopy using a fluorescent intercalant (bar = 8 µm)
Sélection de publication :
- M. Suissa, C. Place, E. Goillot, B. Berge, E. Freyssingeas (2007) “Dynamic light scattering as an investigating tool to study the global internal dynamics of a living cell nucleus” EPL., 78, 3, 38005-38011.
Description of Themes :
Bacteria Swimming in the Vicinity of a Surface
- Lemelle L. (USR3010 & UMR5570 ), Chatre E. (USR 3010), Haftek Z. (USR3010) et Place C. (USR3010 & UMR5672)
- Joint Work With : Palierne J-F (UMR5672) and Vaillant C (UMR5672)
Planktonic bacteria reach mineral or biological surfaces in their environments either through passive transport by convection-diffusion or gravity or, more efficiently, by active propulsion using flagella. On surfaces, the bacterial motility persists in a drastically modified form. Dark field videomicroscopy experiments were developed to statistically quantify this change. A Hydrodynamic model allowed to identify the critical parameters for the bacterial propulsion in the vicinity of surface. Tracking of bacteria in dark-field videomicroscopy at low magnification allows observation of a large number of bacteria under physiological conditions. The protocols for culture of E. coli and for optical imaging techniques in dark field contrast near a planar surface were established. A drop of bacterial cells solution deposited on a glass lamella is observed using an inverse microscope with an EBCCD Hamamatsu camera. This method allows observations of the cells with a strong and constant contrast during the experiment without modifying the dynamics of the cellular swimming (Fig. 1). The trajectories are determined using a data treatment developed at the Joliot-Curie lab, recording the bacterial geometry and their dynamics properties (local velocity, average or median velocity, and radius of curvature).
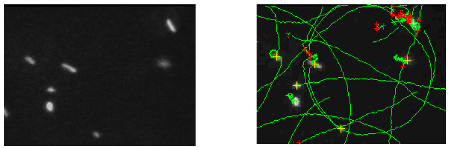
Figure 1a : Dark fiel image of E. coli cell on a glass lamella surface recorded with a x40 objective. Figure 1b : Image of the calculated trajectories (en vert) for the bacteria observed on the previous figure.
Near a solid surface, E. coli becomes trapped and swims in circles. Tracking of bacteria in dark-field videomicroscopy evidences a direct relation between curvature radius of the trajectory and bacterium speed. A hydrodynamic model for this motion on a non-slipping surface was proposed by E. Lauga & al. [Biophys. J. 2006] using the resistive force approximation with moderate agreement with observations. A fair agreement obtains with a revised version of Lauga’s model. Models predict the nanometric distance of the cell to the surface is the critical hydrodynamic parameter. A TIRF LEICA microscope, acquired thanks to a "mi-lourd" support from CNRS, was installed in the Laboratoire Joliot Curie at the end of 2007. Different strategies of physiological fluorescent staining were explored. Finally, bacterial mutants bearing a plasmid that encode for a periplasmic protein fused to mCherry (a red fluorescent protein) were produced and demonstrated the same swimming characteristic as the wild type. Preliminary TIRF videomicroscopy experiments (Figure 2a) with bacteria revealed a nanometric range for the distance of the body to the surface (10 nm < d < 300 nm).
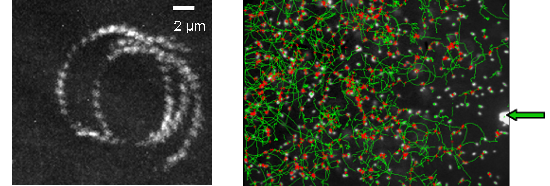
Figure 2a : trajectory of E. coli swimming close to a surface observed by TIRF microscopy (magnification X60, staining Syto). Figure 2b : Trajectories of E. coli (dark field magnification X20) swimming close to a surface while a chemorepellent solution is microinjected on the surface (extremity of the capillary is indicated by a green arrow).
Since hydrodynamics predicts a physical trapping near surfaces, E. coli cells are predicted to be loosely capable to orientate their swimming versus chemicals. Chemo-repellent solutions were microinjected within few m of the surface (Figure 2b). Observations are all consistent with a negative chemotaxis response of cells moving down a spherical repulsive-gradient. The cellular distribution has a radial symmetry centered on the aperture of the capillary. The number of motile cells and their median speed decrease and reach minimal values in less than 100s. Once the capillary is removed, the system recovers its initial state. Responses of the tsr(-) and tar(-) mutants to Ni-microinjection revealed the implication of the MCP receptors. Herein swimming cells can sense surface chemical environments and overcome hydrodynamics laws to move toward preferential environments. Other techniques to induce chemical gradients must be now explored to reach a satisfactory reproducibility.
This work was supported by iterative private subcontracting since 2007 guaranteeing the expenses ( > 20 kEuros) and the salary of an engineer in microbiology.
Two publications are under submition :
- Radius of curvature of E. coli trajectories near a planar surface increases with the swimming speed. by Lemelle L., Palierne J-F., Chatre E. and C. Place.
- Resistive force theory with slip at the boundaries applied to bacterial locomotion in viscous media by Palierne J-F., Lemelle L., Chatre E. and C. Place