Accueil du site > Thèmes de recherche > Previously Hosted Projects > Dynamique d’Assemblages Biologiques - The dynamics of biological molecules
Dynamique d’Assemblages Biologiques - The dynamics of biological molecules
Equipe Argoul
PrésentationThe ‘Dynamics of biological molecules’ team is composed of experimental physicists using micromanipulation and visualization techniques (atomic force microscopy AFM, surface plasmon resonance SPR, high resolution surface plasmon microscopy SSPM, magnetic tweezers …) for single biological object imaging.
Team leader ( Chef d’Equipe ) :
- Françoise ARGOUL - CNRS research director
Team members ( Membres de l’Equipe ) :
- Lotfi BERGUIGA - CNRS research engineer
- Nicolas HUGO – postdoctoral fellow
- Pascale MILANI - postdoctoral fellow
- Fabien MONTEL - PhD student
- Cendrine MOSKALENKO - ENS associate professor
- Thibault ROLAND - PhD student
Several questions have been addressed :
Chromatin remodelling complex actions on nucleosomes : mapping the nucleosome sliding along a DNA fragment. The organization of DNA into nucleosome represents a physical barrier for the transcription factors binding to their target DNA sequences, and interferes with several basic cellular processes. Histone modifications, ATP-remodeling machines and the incorporation of histone variants within chromatin are used by the cell to overcome the nucleosomal barrier and modulate DNA accessibility by the control of nucleosome dynamics. In these experimental studies (conducted in close collaboration with D. Angelov and S. Dimitrov), we use a single molecule technique (Atomic Force Microscopy, AFM) to visualize isolated nucleosomes in order to quantify the influence of histone octamer composition (H2A-Bbd variant) on the equilibrium nucleosome conformation and to map nucleosome mobility induced by a remodeling complex (SWI/SNF, RSC).
What is the role of the DNA sequence on the structure and dynamical properties of chromatin ? This study is directly inspired from the theoretical results obtained by A. Arneodo, B. Audit and C. Vaillant. Their theoretical multi-scale approach of DNA sequences suggests that nucleosome thermal mobility is greatly affected by the existence of long range correlations in the structural disorder of DNA polymer (elastic properties induced by the DNA sequence). We study experimentally how these long-range correlations influence the 2D thermodynamical equilibrium conformation of naked DNA fragments, as well as the formation and diffusion of nucleosomes. The goal of this project is to elucidate the link between the DNA sequence and the structural and mechanical properties of the chromatin fibre for a better understanding of functional genome organization. Two experimental methods are applied to provide this experimental demonstration : AFM imaging of DNA and nucleosomes adsorbed onto mica in low ionic strength solutions, and SSPM on gold adsorbed nucleosomal particles in air and in liquid media.
Imaging nanoscale objects by a nonintrusive optical method. In parallel to more conventional SPR and AFM studies, a high resolution surface plasmon microscope has been constructed in LJC that allows the imaging of nanoscale particles (both metal and polymer) down to 5 nm in size. The advantage of this optical method and its complementary interest with respect to other methods rely on two aspects, (i) it is non intrusive and non destructive since it does not require any fluorescent dye tagging, (ii) it is very versatile because it can be run in liquid media and coupled to microfluidic systems.
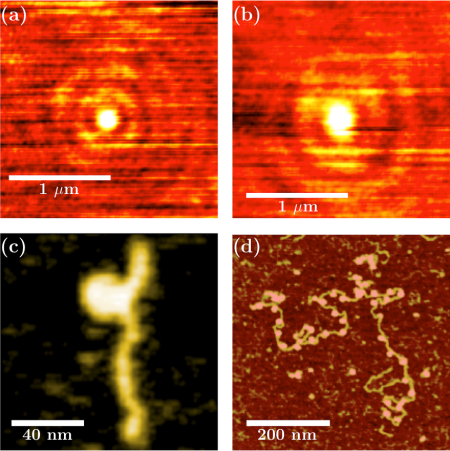
(a) SSPM image of a 30nm latex nanoparticle in water (V(z) function is coded from 0 to its maximum corresponding to the center of the particle).
(b) SSPM image of a mononucleosome in air, we notice in this figure that the Point Spread Function is greater in air ( 200 nm) than in water ( 150 nm).
(c) AFM topographic image obtained in tapping mode in water of a mononucleosome reconstituted on a genomic sequence extracted from the yeast Saccharomyces cerevisiae genomic DNA (389 bp). Color scale 0 to 5 nm.
(d) AFM topographic image obtained in Tapping Mode in air, for chromatin reconstituted on 33 repetitions of 601 positioning DNA sequence (repeat length 197 bp) with the histone variant H2A.Bbd. Color scale : 0 to 2 nm
Sélection de publications :
- L. Berguiga, S.J. Zhang, F. Argoul, J Elezgaray, High resolution surface-plasmon imaging in air and in water : V(z) curve and operating conditions, Optics Lett. 32 (2007) 509-511.
- L. Berguiga et F. Argoul : "Microscope de plasmon de surface haute résolution à balayage en interférométrie hétérodyne avec polarisation radiale” patent : ref CNRS DI 01877-01 (depot le 11/12/2007)
- F. Montel, E. Fontaine, P. St-Jean, M. Castelnovo, C. Faivre-Moskalenko, AFM imaging of SWI/SNF action : mapping mononucleosome remodeling and sliding, Biophys. J. 93, 566-578 (2007).
- J. Moukhtar, E. Fontaine, C. Faivre-Moskalenko, A. Arneodo Probing persistence in DNA curvature properties with Atomic Force Microscopy, Phys. Rev. Lett. 98 178101-1 (2007).