Biophysique
Les échanges entre les disciplines ont depuis toujours conduit à changements de paradigmes et à des découvertes majeures dans le domaine scientifique. Ces dernières années, les interactions accrues entre biologie et sciences formelles comme les mathématiques, l'informatique ou la physique ont ainsi donné des résultats spectaculaires.
Au LBMC, nous sommes convaincus que l'intégration de ces disciplines représente une stratégie de recherche essentielle en biologie et nous voulons promouvoir cette approche dans nos projets.
En particulier, nous pensons que la (bio)physique a un rôle majeur à jouer dans cette stratégie interdisciplinaire. De nombreuses approches biophysiques sont désormais systématiquement intégrées dans nos programmes de recherche :
-
D’un point de vue "pratique", via l’utilisation d’instruments expérimentaux développés par la communauté (bio)physique, tels que des dispositifs microfluidiques ou optiques, parfaitement adaptés à l'étude des systèmes vivants ou des biomolécules.
-
D'un point de vue plus "conceptuel", via le développement des modèles biophysiques permettant de comprendre comment les fonctions de processus biologiques dépendent de leurs propriétés mécaniques et/ou thermodynamiques.
Ainsi au fil des années, le LBMC a su développer un réseau de collaboration avec la communauté biophysique, notamment localement avec le Laboratoire de Physique de l’ENS de Lyon, afin de faire fructifier ses recherches.
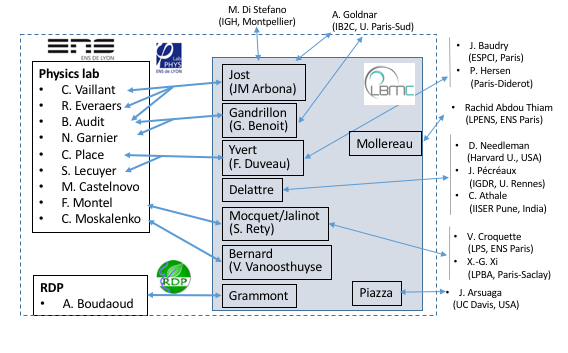
Aperçu des interactions entre équipes du LBMC et biophysicien(ne)s en 2021
Exemples de projets interdisciplinaires actuels en biophysique (*:biophysicists, +:biologists)
Physical Biology of Chromatin
The team Physical Biology of Chromatin (D. Jost) at LBMC is mainly composed of biophysicists. In close connection with other theoreticians and experimentalists, our research addresses generic or specific biological questions on chromatin and gene regulation by developing physical and computational approaches. In particular, we focus on:
- The coupling between genome 3D organization and functions in collaboration with C. Vaillant* [LPENSL] M. Di Stefano*, G. Cavalli+ [IGH, Montpellier], K. Bystricky+ [LBME, Toulouse], and the groups of G. Yvert+, B. Loppin+, O. Gandrillon+ & P. Bernard+ at LBMC.
-
The mechanisms of replication initiation in collaboration with B. Audit* & P. Borgnat* [LPENSL], A. Goldnar* [IB2C, Paris] & the group of O. Hyrien+ [ENS, Paris].
Polymer modeling of the chromosome reorganization at the histone-to-protamine transition during cricket spermiogenesis (collaboration with Loppin’s team [LBMC]).
Evolution & conservation of spindle positioning in nematodes
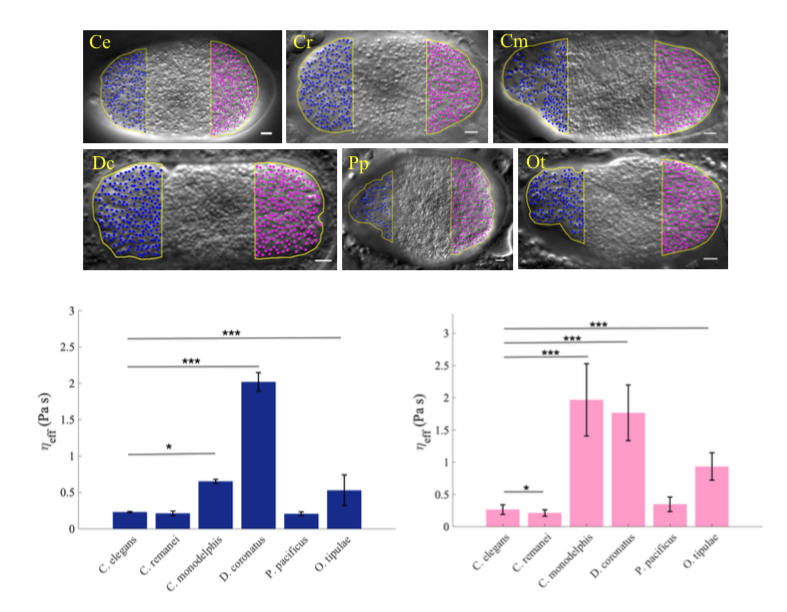
This project is a collaboration between Delattre+'s team and the group of Dr Chaitanya Athale* at IISER Pune, India. The remarkable conservation in basic cell functions across organisms raises a vital question: to what extent do cellular mechanisms evolve without disrupting the basic function that they sustain? Our team quantified spindle positioning during the first cell division of 42 nematode species and found rapid changes in the underlying mechanisms, despite conservation of asymmetric positioning. Our goal now is to identify physical and molecular parameters that act to either constrain change or enforce flexibility and innovation across species.
In collaboration with the biophysics group of C. Athale, we aim to i) measure key biophysical parameters, ii) extend pre-existing mechanical models of the C. elegans spindle to explore parameter changes between species, iii) experimentally validate evolutionary predictions.
Cytoplasmic viscosity measured in 6 different nematode species, from diffusion coefficient of cytoplasmic granules. Blue and pink are anterior and posterior cellular poles, respectively.
LITREA: Deciphering nuclear receptor biological function in differentiating mouse pluripotent embryonic cells
This is a project headed by G. Benoit+ (SBDM team, LBMC) and in collaboration with P. Annibale* (MDC Berlin), B. Audit* (LPENSL), A. Goldar* (CEA) and J.-M. Arbona* & D. Jost* (LBMC). It aims at deciphering the dynamic interplay between specific transcription factors belonging to the nuclear receptor superfamily and its functional consequences in shaping the transcriptional response and biological outcome along retinoic acid-induced pluripotent embryonic cells differentiation. For this it will combine state of the art genome modification methods and fluorescent microscopy techniques allowing single molecule microscopy of TF binding and transcription kinetics with computational method of signal analysis and dynamic modelling.
Light scattering for probing nuclear dynamics
This is collaboration between G. Benoit+ (SBDM team, LBMC) and in collaboration with the team of E. Freyssingeas* at the LPENSL. This team developed an original light scattering experimental device that allows the assessment of the global internal dynamics of the nucleus of a living cell along important biologically relevant processes (e.g. cell cycle, differentiation). This device will be used to probe the nuclear organisation dynamics and its changes along mES/EC cells differentiation.
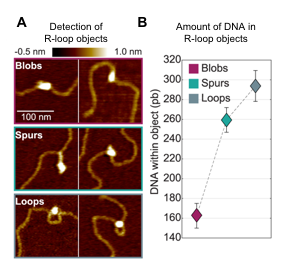
3D organisation of R-loops
This is a collaboration between V. Vanoosthuyse+ [LBMC, Team Bernard] and C. Moskalenko* & F. Montel* [LPENSL]. R-loops are three-stranded transcription byproducts that form when the nascent RNA unduly hybridizes with its template strand. Recent evidence suggests that a subset of R-loops compromises genome integrity, particularly during DNA replication. The characteristics of such toxic R-loops remain poorly defined. Using single molecule approaches in vitro, we have shown previously that R-loops differ by their 3D organisation and by the physical constraints that they impose on the surrounding DNA. We are currently trying to understand what determines this 3D organisation and whether it could contribute to the cytotoxicity of R-loops.
Detection of R-loop dependent secondary structures (“R-loop objects”) using AFM. A. Type of objects. B. Amount of DNA in each type of objects.
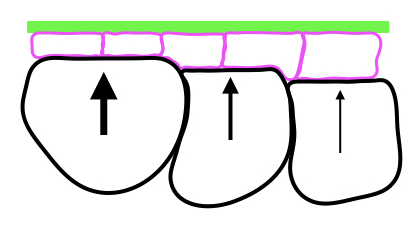
Mechanics of epithelial cells in Drosophila
This is a collaboration between M. Grammont+ (LBMC) and A. Boudaoud* (Ecole Polytechnique). Epithelial cell shape depends on the intracellular actomyosin activity and on the surrounding tissues that may push or pull the epithelial cells. We aim to describe, quantify and disturb these extra-cellular forces and constraints in order to understand their impact on epithelial cell morphogenesis. To reach this aim, we use interdisciplinary tools, such as genetics, live imaging, 3D reconstruction, atomic force microscopy.
Because of the growth of internal cells (black) and of the stiffness of the basement membrane (green), the epithelial cells (pink) are forced to change shape (from cuboidal to squamous).
Structure, function and mechanism of helicases
The PRIO team (P. Jalinot+ & V. Mocquet+) has several collaborations with biophysics groups for studying molecular mechanism of helicases at the single molecule level. Helicases are enzymes which can unwind nucleic acids structures folded as double strand, hairpin or G-quadruplex. Unfolding of nucleic acid can be easily monitored by single molecule techniques as magnetic tweezers (collaboration with V. Croquette*, LPS ENS Paris), smFRET (collaboration with X.G. Xi*, LBPA, ENS Cachan) and optical tweezers (collaboration with F. Montel*, Physics Lab ENS Lyon). This latter collaboration has been started in order to develop at ENS Lyon an instrument combining in the same apparatus two single molecule techniques (optical tweezers and smFRET) in order to monitor in real time nucleic acid unfolding and helicase conformational changes.
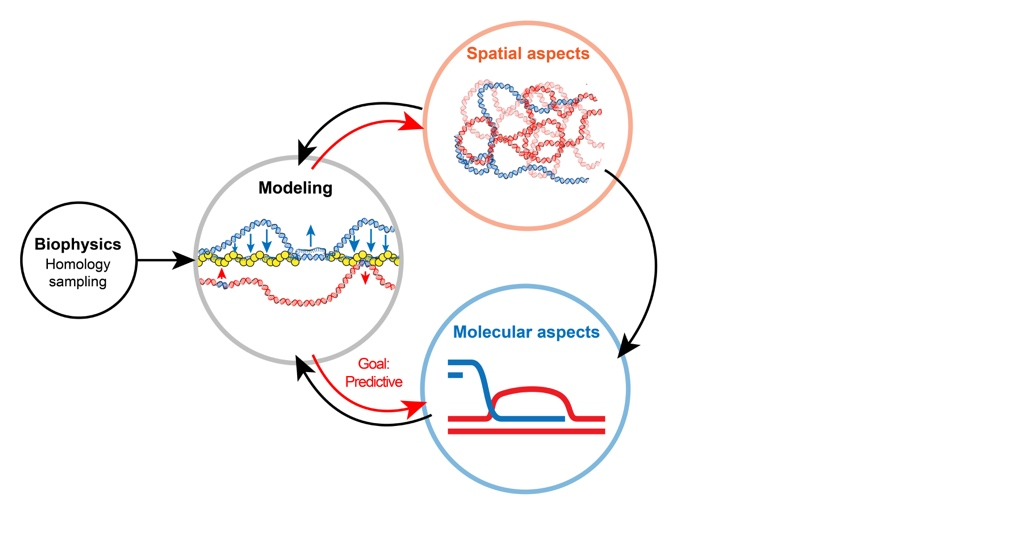
Interplay between DNA break repair and spatial chromatin
DNA break repair by homologous recombination entails the identification by the broken region of an intact, identical DNA molecule in the genome and nucleus. Homology search thus minimally requires a machinery capable of sampling dsDNA for a match (molecular scale) and opportunities for spatial collision with a homologous donor (cytological scale). The Piazza+’s group studies this multi-scale process by coupling diverse proximity ligation-based assays to track both chromatin folding and site-specific recombination progression in S. cerevisiae (Collaboration with R. Koszul*, Institut Pasteur). We are developing stochastic simulations of the search process to attempt determining more quantitatively the contribution of various nucleoprotein factors whose precise role remain elusive (collaboration with J. Arsuaga*, UC Davis).
An interdisciplinary approach to tackle homologous repair

![Fig.2: Polymer modeling of the chromosome reorganization at the histone-to-protamine transition during cricket spermiogenesis (collaboration with Loppin’s team [LBMC]).](https://www.ens-lyon.fr/LBMC/images/activites-du-laboratoire/interdisciplinarite/biophysique/fig-2-polymer-modeling-of-the-chromosome-reorganization-at-the-histone-to-protamine-transition-during-cricket-spermiogenesis-collaboration-with-loppin2019s-team-lbmc)