Biomath
A l'instar d'autres disciplines scientifiques contemporaines, la biologie moléculaire et cellulaire soulève des défis mathématiques et informatiques nouveaux qui sont inhérents à la production massive de jeux de données hautement complexes et multi-dimensionnels. Cette complexité toujours croissante implique le développement d'une recherche interdisciplinaire de pointe. Le LBMC met l'accent sur ce type d'approche interdisciplinaire via le développement de collaborations scientifiques avec des mathématicien(ne)s, des bioinformaticien(ne)s et des statisticien(ne)s qui ont l'opportunité de développer leur propre méthodologie au sein du laboratoire et de s'attaquer aux défis actuels de la biologie computationnelle.
La Biologie des Systèmes est un axe de recherche majeur du LBMC. Son objectif est d'atteindre une compréhension quantitative de la dynamique des processus biologiques par l'utilisation d'analyses mathématiques et statistiques qui intègrent les données biologiques, ceci afin de développer des modèles prédictifs des comportements biologiques. Dans cette optique, nous développons actuellement notre recherche autour de deux lignes méthodologiques : La modélisation dynamique (Dynamical Modelling) et l'IA et l'apprentissage automatique (AI & Machine Learning).
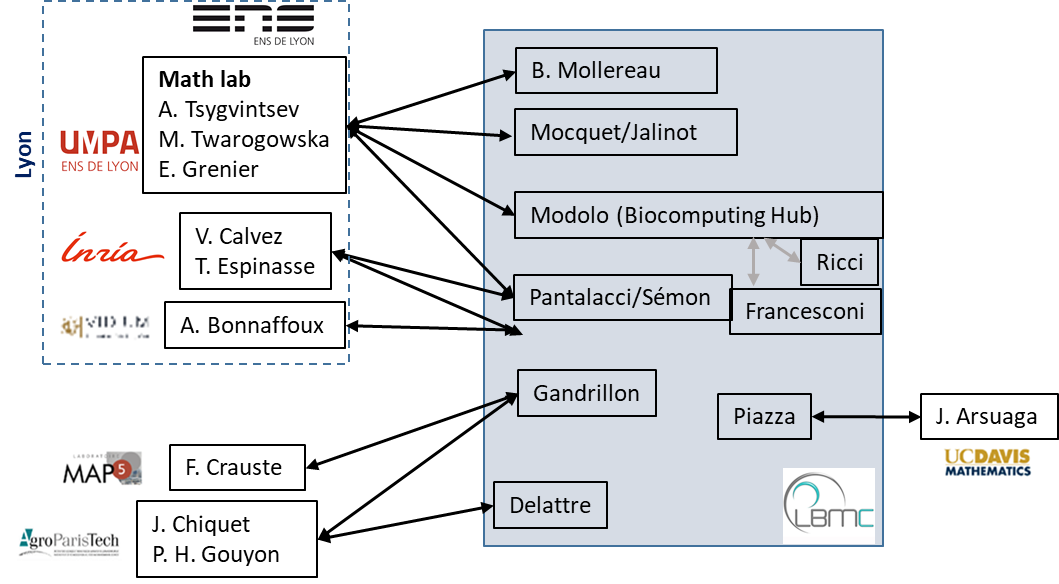
Aperçu des interactions entre équipes du LBMC et mathématiciens (2021).
L'environnement Math-Bio-Info du LBMC est décrit plus amplement ici.
Modélisation dynamique (dynamical modeling)
Les approches centrées sur un gène, couramment utilisées au cours des dernières décennies, représentent toujours un excellent moyen de concentrer la puissance expérimental et le raisonnement scientifique sur des mécanismes spécifiques. Cependant, ce type d'approche doit être complété par une perspective plus large qui intègre la complexité du contexte biologique. A l'inverse, une compréhension fine des processus cellulaires dans toute leur complexité n'est pas réaliste, et souvent superflue.
Le défi à venir est donc d'extraire des principes généraux, possiblement des règles simples, qui régissent les propriétés des cellules vivantes, tout en embrassant la multiplicité et complexité des processus moléculaires sous-jacents. Ceci est possible au niveau théorique, ce qui peut permettre de développer de nouveaux concepts. Néanmoins, toute théorie doit être confrontée à la réalité des systèmes biologiques. Pour cela, nous avons besoin :
- De données expérimentales quantitatives qui sont soit spécifiques, soit suffisamment volumineuses pour rendre compte des nombreuses molécules impliquée. Idéalement, ces données doivent offrir une haute résolution dans le temps et dans l'espace.
- De modèles mathématiques et informatiques qui représentent et étudient les propriétés dynamiques et collectives des entités d'intérêt (molécules, cellules...).
- D'interactions régulières entre les prédictions des modèles et les résultats expérimentaux pour obtenir des conclusions robustes et rigoureuses.
Le développement de ce type de modèles mathématiques ou computationnels est mis en place par plusieurs groupes de recherche du LBMC comme décrit ci-dessous.
Systems Biology of Decision Making - O. Gandrillon
The inference of dynamical gene regulatory networks (GRNs) is a long standing theoretical and computational challenge that we tackle with very original approaches like optimal transport and stochastic processes. Once the GRNs have been inferred another challenge is to connect the cellular behavior to the behavior of the inner GRNs thanks to a dynamical PDE model incorporating molecular variables, or multiscale agent-based models.
This line of research will be developed through the Mémoire ANR project that involves collaboration with the Dracula team (INRIA), the ICL team (CIRI), Fabien Crauste (Université de Paris) and two private companies, Altrabio and Vidium.
Physical Biology of Chromatin - D. Jost
Epigenetic markers are tightly coupled to gene expression and cell lineage. Our group studies:
- The spreading of epigenetic marks coupled to three dimensional structure of the genome using mechanistic dynamical models, with a focus on silencing marks and DNA repair.
- The relationship between origins of replication and epigenetic landscape using mechanistic single cell models to reproduce experimental characterisation of the cell such as Mean Replication Timing using epigenetic marks, in collaboration with Benjamin Audit (ENS de Lyon) and Olivier Hyrien (ENS Paris).
Comparative and Integrative Genomics of Organ Development - S. Pantalacci / M. Semon
In the team, we focus on the temporal dynamics of developmental mechanisms as a key determinant of their outcome. In collaboration with Monika Twarogowska (UMPA, ENS de Lyon) and Vincent Calvez (Institut Camille Jordan, Univ Lyon 1), we have embedded a classical Turing model in an original framework and shown this was sufficient to recapitulate the complex dynamics of tooth patterning and predict its perturbation. We will now infer dynamical GRN for this patterning mechanism and study both conceptually and experimentally its variational properties throughout evolution.
Genome mechanics - A. Piazza
Our work aims at understanding the interplay between the spatial organization of the genome and the DNA repair process by homologous recombination. In particular, we focus on the homology search step, in which the broken DNA molecule samples genomic DNA for a homologous match. The stringency of this dynamic process promotes genomic stability, and involves multiple layers of control, at the molecular and chromosomal levels. To gain insights into this rate-limiting step of the repair process we develop various quantitative or semi-quantitative assays that provide kinetics information on the repair process, coupled with Hi-C to determine the averaged pair-wise dsDNA contact frequency genome-wide.
In collaboration with the laboratory of Javier Arsuaga (Department of Mathematics, UC Davis), we attempt to mathematically model the repair process in order to derive the quantitative contributions of multiple nucleoprotein factors involved in the search process, at the local level (e.g. recombination proteins) and chromosomal level (e.g. chromatin folding by cohesins).
Apoptosis and Neurogenetics - B. Mollereau
Most cancer treatments, such as chemotherapy or radiation-based therapy, eliminate cancer cells by inducing apoptosis, which is the most well studied form of programmed cell death. Although these treatments proved relatively successful for several cancers, malignant cells often develop resistance to apoptosis leading to the reduction or loss of treatment efficiency. There is thus an urgent need for alternative treatment for cancer therapy. Our consortium has found that programmed necrosis, which has been neglected for many years, is a potent inducer of cell death in cells that are resistant to apoptosis.
Previous results obtained by our consortium revealed an intriguing finding: blocking the activation of caspases, the molecular scissors that actually demolish the cell during apoptosis, enhances programmed necrosis in both fly retina cells and mice testis. Corroborated with literature reports suggesting that cancers could use non-lethal caspase activation in their own advantage, we propose and will investigate whether caspases could in fact block programmed necrosis in several malignancies such as melanoma, glioblastoma, lung and pancreatic cancers.
To understand the switch from apoptosis to necrosis in cancer cells, we will characterize the dynamic of the inhibition of necrosis by caspases by performing live imaging and mathematical modelling with Emmanuel Grenier (UMPA, ENS de Lyon).
RNA metabolism in immunity and infection (RMI2) - E. Ricci
In the team, we are interested in studying the dynamics of messenger RNA (mRNA) translation by ribosomes and its cross-talk with other cellular processes such as mRNA degradation. mRNA translation is a tightly regulated process in which ribosomes decode the information contained in the open reading frame (ORF) of mRNAs into a polypeptide sequence. Ribosome recruitment onto the mRNA and its translocation across the ORF are not a uniform process. Various obstacles can affect the progression of ribosomes and modify their decoding speed leading to conflicting situations such as ribosome stalling or collisions that can induce the degradation of the mRNA and the nascent polypeptide.
We use ribosome profiling coupled to next-generation sequencing to map the position of ribosomes on the coding sequence of all expressed mRNAs under regular conditions and after viral infection or immune cell activation. Using this data we then try to develop ODE-based mathematical models to explain ribosome progression across open reading frames based on transcript cis-acting features such as codon usage, GC content and length of untranslated regions.
Genetic complexity of living systems - G. Yvert
Genetics - the science that links phenotypes to genotypes - is rarely adapted to the highly-dynamic nature of living systems. For example, a classical approach is to introduce a mutation in a model organism and then examine the resulting phenotypic change. This can demonstrate the link but rarely its mode of action: how is this mutation causing this effect?
We are developing novel methods and concepts to understand how mutations modify the dynamics of molecular regulations. For example, by coupling experiments with mathematical modelling of gene regulations, we were able to delineate an intermediate layer of the genotype-phenotype association: specific model parameters had to be re-adjusted in response to a mutation, highlighting the molecular path by which this mutation operated.
Another approach is to study how natural selection and evolution shapes the adaptation of organisms to dynamic environments, which indicates the mutations that modify dynamic regulations in a beneficial way. We are also implementing optogenetic technologies enabling genetic perturbations with extreme spatiotemporal precision. This allows us to monitor the dynamic effects of mutations in real time. Coupling such experiments with kinetic models of stochastic processes can reveal how cells and organisms adapt - or not - to de novo mutations.
IA et apprentissage automatique (AI & Machine Learning)
Les méthodes d'analyse de données conventionnelles sont principalement conçues pour des jeux de données de faibles dimensions et ont besoin d'être adaptées à des données plus complexes et à plus de dimensions. Par exemple, de nouvelles méthodes de machine learning doivent être développées afin de représenter, d'analyser, de synthétiser, d'intégrer et d'exploiter tout le potentiel des données de génomique de cellules uniques.
En effet, le développement récent de méthodes basées sur les technologies d'analyse de cellules uniques offrent la possibilité de mesurer à l'échelle génomique des propriétés de l'ADN, de l'ARN, de l'état chromatinien, des protéines ou des métabolites d'un grand nombre de cellules individuelles. Ces techniques nous permettent d'étudier la variabilité de cellule à cellule au sein d'un échantillon biologique et d'aborder de nouvelles questions au delà des limites de la génomique classique.
Cette complexité a atteint un niveau sans précédent avec le développement d'approches de transcriptomique à résolution spatiale (spatial transcriptomics) qui connectent les données moléculaires à la localisation cellulaire de paramètres multi-dimensionnels. Relever ces défis requiert le développement de méthodologies complexes de machine learning basées sur des approches non-paramétriques et multi-échelles.
Ce type de recherche est actuellement poursuivi par les équipes du LBMC décrites ci-dessous.
Systems Biology of Decision Making - O. Gandrillon
Investigating cell-to-cell variability is fundamental to better understand differentiation, as it reflects the intrinsic stochastic molecular processes and provides information on the underlying molecular networks at play. This line of research will be developed through the SingleStatOmics ANR project that gathers a national interdisciplinary consortium in machine learning / AI dedicated to single cell genomics, with experts in machine learning, optimal transport and statistics, both from the academic (Bertrand Michel, EC Nantes, Filippo Santambrogio, UCBL, Julien Chiquet, INRAE) and the private sector (Jean-Philippe Vert, GoogleBrain).
Physical Biology of Chromatin - D. Jost
To study the replication process at the single cell level, we developed a deep-learning based algorithm to decode the electrical current signals that output from nanopore sequencing into a DNA sequence with modified nucleotides. These nucleotides are incorporated in the cell during the replication process, allowing to monitor, genome-wide in yeast, the direction and speed of the replication forks at the single cell level. We are currently developing a method to monitor the timing of replication at the single cell level, in collaboration with Benjamin Audit (ENS de Lyon) and Olivier Hyrien (ENS Paris).
Quantitative regulatory genomics - M. Francesconi
In Francesconi’s team we are interested in understanding genetic and non-genetic sources of phenotypic variability. We focus on genome-wide gene expression dynamics – both at single-cell and single-individual level - as a multidimensional intermediate phenotype to infer information higher level phenotypes such as cellular states developmental stage. For example, using big data integration and machine learning approaches we recently developed a computational method to accurately infer developmental age of single individual expression profiles which we are now extending to single cell expression.
Post-transcriptional Regulation in Infection and Oncogenesis - Jalinot/Mocquet
We have developed an experimental approach aiming at identifying small chemical compounds prone to induce specifically proteolytic degradation of a given protein. The method is based on the analysis of the ratio between the fluorescences of a hybrid protein associating a protein of interest to a first fluorescent protein and a second control fluorescent protein. The measure is performed by flow cytometry analysis of cells expressing the two proteins. As the effect exerted by unoptimized compound can be faint, we have initiated a collaboration with Alexei Tsygvintsev (UMPA, ENS de Lyon) who develops artificial intelligence approaches for the analysis of experimental biology data to see if elaboration of an AI method using either graphical or numerical data sets can help to spot interesting candidates in the first screening round.
Comparative and Integrative Genomics of Organ Development - S.Pantalacci/M. Semon
In collaboration with mathematician Laurent Guéguen (LBBE, Univ Lyon 1) and biostatistician Marie-Laure Delignette-Muller (VetAgro-Sup, Univ Lyon 1) we model quantitative data, including RNA-seq data and morphological measurements, to extract temporal dynamics and compare them across species.
Scientific environment / Partners
The ENS-Lyon campus has long been involved in the developments of math-info-bio interactions, with the Complex Systems Institute (IXXI), that promotes modeling research in applied science, with a special focus on AI challenges in Systems Biology for the 2021-2026 mandate.
In particular, systems biology has been very active in Lyon, especially thanks to the Lyon federation for Systems Biology (BioSyl), and to the structuring effect of INRIA groups in the Lyon Campus (Virtual plant, Dracula, Numed, Bamboo, Beagle).
In addition, the Scidolyse consortium has emerged, dedicated to research in machine learning in Lyon Saint Etienne, with founding members being part of the UMPA, LIP and LBMC labs (among others).
The LBMC has also long been dedicated to interdisciplinary research (bio - math - physics) by developing strong connexions with INRIA and the Camille Jordan Institute (math lab, UCBL).
The recent funding of the Spatial-Cell-Id Equipex project will be a superb occasion to foster interaction between the LBMC and the Spatial-Cell-Id core units (IGFL, RDP ad SBRI).