PLA
CONTEXT:
This program has been developped and published in the context of a scientific project of the team of Dr Didier Auboeuf.
The PLA macro for the program ImageJ is based on a previously released macro (NUKE-BREAK) and optimized to detect Proximity Ligation Assay foci within nuclei of human cells. The macro was designed to automatically detect and process batches of microscope coupled acquisitions. To this end, key parameters for nuclei and foci detection (minimum and maximum surface and circularity) have been optimized.
BRIEF DESCRIPTION:
The PLA macro explores recursively a specific folder and treat all images with the correct extension (chosen by the user).
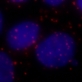 |
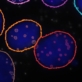 |
| Original | Treated |
The macro will generate a listing of couples of acquisition images using user defined DAPI.extension and PLA.extension.
Using the GUI, the user can specify the extensions as the main analysis parameters:
- Minimum Surface for initial nuclei detection
- Maximum Surface for initial nuclei detection
- Maximum Surface of a single nucleus (for nuclei aggregates identification and splitting)
- Minimum Circularity of single nucleus (for nuclei aggregates identification and splitting)
- Minimum Surface for PLA foci
- Maximum Surface for PLA foci
These parameters can be changed and saved, so the program can be adapted to specific acquisition resolution and/or experimental conditions affecting the geometry of the nuclei and the PLA foci.
The images couples are then processed as follows:
- The noise of both DAPI and PLA images is independently removed using the “substract background” function.
- The DAPI channel is then used to detect the Nuclei with the "Huang" thresholding method and the initial Nuclei Minimum surface and Maximum surface parameters.
- The aggregates of nuclei are subsequently identified using the single Nucleus Maximum surface and Minimum Circularity. Detected ROIs fitting with these parameters are then splitted using the "Watershed" algorithm.
- PLA foci are then detected independently for every isolated nucleus, using the “Max-Entropy” threshold method and the foci Minimum Surface and Maximum Surface parameters.
- Finally, the program collects the size of all nuclei and foci to generate a Results.csv table.
DOWNLOAD:
