Spatial genome organization and DNA repair
Homologous recombination (HR) is an elaborate DNA break repair pathway that uniquely exploits an intact homologous double-strand DNA (dsDNA) molecule as a template for repair. The search for a homologous molecule, which can be either the sister chromatid, the homologous chromosome, or dispersed repeats, is subdued to the initial and evolving collision probabilities between the break site and other genomic loci over the course of the repair. We investigate how factors that spatially organize chromatin in the nucleus impinge on the efficiency and accuracy of HR over time. To this end, we use and develop custom high-throughput contact genomics approaches and assays for physical detection of HR intermediates in the model eukaryote S. cerevisiae in which a single site-specific DNA break can be rapidly induced in almost all cells of a population.
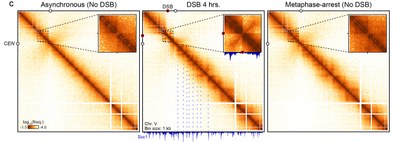
Figure 1: Hi-C map of chromosome 5 before (left) and after (center) formation of a DNA break (red dot), showing global and local changes to chromatin contact frequencies. Cohesin enrichment sites correspond to loop pattern along the chromosome, highly similar to that of metaphase-arrested, undamaged cells (right).
For instance, we recently showed that HR repair takes place in the highly organized context of metaphase chromosomes, in which chromatin is structured as arrays of loops by cohesins (Figure 1). An independent set of factors maintain spatial proximity of the DSB ends and act as a roadblock for cohesin. This interaction leads to the early recruitment of the break site at the chromosome axis and inhibits interaction with other chromosomes. Kinetics study of the repair process with competing intra- and inter-chromosomal region of homologies enabled us to address the biological consequence of this organization: to bias repair away from inter-chromosomal donors (Figure 2). We pursue the investigation of these and other factors in regulating the homology search process and its impact on genome stability.

Figure 2: Model of homology search regulation by cohesin-mediated chromatin folding
