Publication from the IGFL in Current Biology on April 1st, 2019.
 The genome, which corresponds to the whole DNA content of a cell and is characteristic of an organism, can present big variations in size between species. If this size does not correlate with organismal "complexity", it is not clearly established yet if bigger or smaller genomes could be adaptive. Several mechanisms can impact genome size : duplications, deletions, and multiplication of particular sequences called transposable elements. These sequences, which can insert and propagate in genomes, are mostly neutral for the host, or sometimes deleterious if they insert into or next to a gene. They represent at least 45 % of the human genome. Correlations between transposable element coverage and genome size have already been observed in animals or plants, involving large transposable elements described as "autonomous" since they encode the proteins needed for transposition.
The genome, which corresponds to the whole DNA content of a cell and is characteristic of an organism, can present big variations in size between species. If this size does not correlate with organismal "complexity", it is not clearly established yet if bigger or smaller genomes could be adaptive. Several mechanisms can impact genome size : duplications, deletions, and multiplication of particular sequences called transposable elements. These sequences, which can insert and propagate in genomes, are mostly neutral for the host, or sometimes deleterious if they insert into or next to a gene. They represent at least 45 % of the human genome. Correlations between transposable element coverage and genome size have already been observed in animals or plants, involving large transposable elements described as "autonomous" since they encode the proteins needed for transposition.
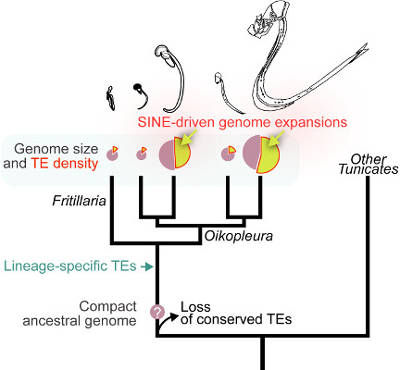
Naville, Henriet et al, within the frame of a collaboration between the Institut de Génomique Fonctionnelle de Lyon (Jean-Nicolas Volff team) and the Sars International Centre for Marine Molecular Biology of Bergen, Norway, studied the genomes of Larvaceans, which are tunicates forming a large portion of the zooplankton. These species harbor small genomes in comparison to other animals, but these genomes present a 12-fold variation in size within the group, this size being correlated to the size of the organism. In an article published in the review “Current Biology”, the researchers could show that these important size variations are mainly due to variable quantities of a particular type of transposable elements called SINEs. SINEs are short elements (a few hundred base pairs) that do not encode any proteins, but rather use the enzymes encoded by bigger, related transposable elements to ensure their transposition ; they can thus be considered as "parasites of parasites".
Such an impact of short non-autonomous transposons on genome size had never been observed before. This work thus reveals a new mechanism of genome expansion linked to fluctuations in the size of organisms. Further studies on these planktonic species will probably allow to better understand the adaptive significance of genome size variation.
Massive Changes of Genome Size Driven by Expansions of Non-autonomous Transposable Elements. Naville M, Henriet S, Warren I, Sumic S, Reeve M, Volff JN, Chourrout D. Curr Biol. 2019 Apr 1;29(7):1161-1168.e6. doi: 10.1016/j.cub.2019.01.080.






