Publication of the IGFL in the journal PNAS on July 1st, 2022. CNRS-INSB communication on July 12, 2022.
Some organisms have the fascinating capacity to regenerate lost body parts. To which extent regeneration entails the redeployment of an embryonic developmental program is a long-standing question of regenerative studies, with implications for development, evolution, and regenerative medicine. In this study, we address this question by comparing the global transcriptional dynamics of leg regeneration and leg development in the crustacean Parhyale hawaiensis. We show that despite extensive overlaps in gene usage, the development and regeneration of Parhyale legs show distinct temporal profiles of gene expression that cannot be aligned in a coherent fashion. These results suggest that regeneration does not simply mirror development, but deploys some of the same gene modules in a different overall framework.
Regenerating animals have the ability to reproduce body parts that were originally made in the embryo and subsequently lost due to injury. Understanding whether regeneration mirrors development is an open question in most regenerative species. Here, we take a transcriptomics approach to examine whether leg regeneration shows similar temporal patterns of gene expression as leg development in the embryo, in the crustacean Parhyale hawaiensis. We find that leg development in the embryo shows stereotypic temporal patterns of gene expression. In contrast, the dynamics of gene expression during leg regeneration show a higher degree of variation related to the physiology of individual animals. A major driver of this variation is the molting cycle. We dissect the transcriptional signals of individual physiology and regeneration to obtain clearer temporal signals marking distinct phases of leg regeneration. Comparing the transcriptional dynamics of development and regeneration we find that, although the two processes use similar sets of genes, the temporal patterns in which these genes are deployed are different and cannot be systematically aligned.
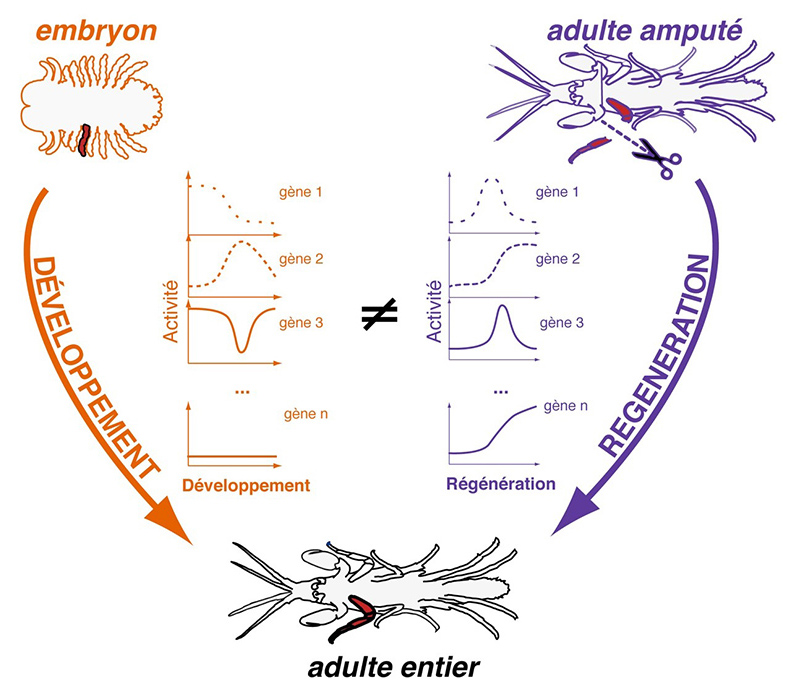
Illustration credits: Michalis Averof
Figure credits: Mathilde Paris
Reference: Distinct gene expression dynamics in developing and regenerating crustacean limbs.. Chiara Sinigaglia, Alba Almazàn, Marie Lebel, Marie Sémon, Benjamin Gillet, Sandrine Hughes, Eric Edsinger, Michalis Averof and Mathilde Paris. PNAS, July 1st, 2022.
DOI : https://doi.org/10.1073/pnas.2119297119






