Publication of the LBMC in the journal Nature Methods on July 11, 2022. CNRS-INSB communication on July 12, 2022.
Transcriptomic data is often affected by uncontrolled variation among samples that can obscure and confound the effects of interest. This variation is frequently due to unintended differences in developmental stages between samples. The transcriptome itself can be used to estimate developmental progression, but existing methods require many samples and do not estimate a specimen’s real age. Here we present real-age prediction from transcriptome staging on reference (RAPToR), a computational method that precisely estimates the real age of a sample from its transcriptome, exploiting existing time-series data as reference. RAPToR works with whole animal, dissected tissue and single-cell data for the most common animal models, humans and even for non-model organisms lacking reference data. We show that RAPToR can be used to remove age as a confounding factor and allow recovery of a signal of interest in differential expression analysis. RAPToR will be especially useful in large-scale single-organism profiling because it eliminates the need for accurate staging or synchronisation before profiling.
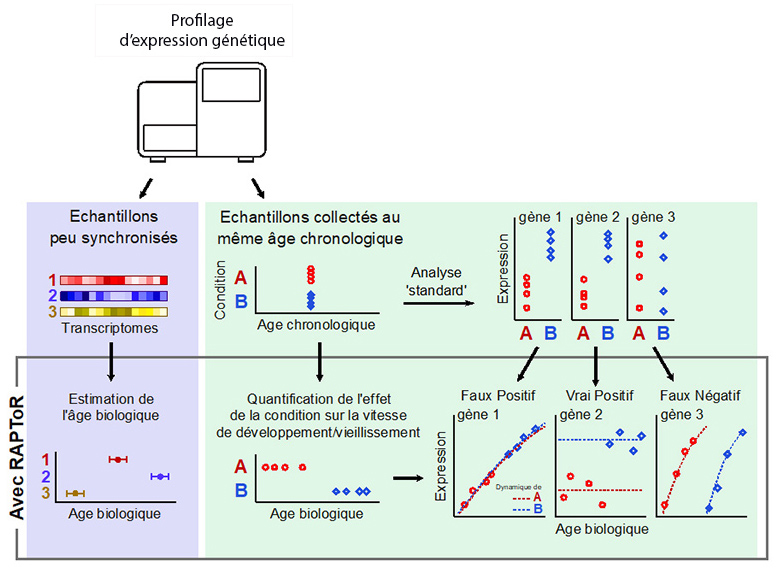
Illustration and figure credits: Mirko Francesconi
Reference: Real age prediction from the transcriptome with RAPToR.. Bulteau R, Francesconi M. Nature Methods, July 11, 2022.
DOI : http://dx.doi.org/10.1038/s41592-022-01540-0






